Gene
KWMTBOMO02602 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002629
Annotation
odorant_binding_protein_LOC100301497_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.885
Sequence
CDS
ATGACGTCCAAAGTATTATTGTCCTGCGTGGTTCTCGCAGTGTTGGCTACTACAGTTTTAGCAGAGGATTCCAGGAAACTCGTCAGTTTCGCCCCGGAGGTGGCTAAAAAATTGAAAGTCCTGATTCAGGAGTGTTTGAATGAAAACGGCTTGGGTGAAGACGCGATAGAAGTAATACGTGCCGGTGAATATAGAGAGGACGAACCATTCCAGAACTTAGTTTATTGCGCTTATAAAAAATTCGGAGCGCTCGATGAAAATAATCGAATCATCAGTCAAGTTGCAGCTGCGTCTTTCCCGAAAGACATAGACGTGGTCACTGTTATCGAGAGCTGCGGAAAAGAAGATGGAAATACCCCAGTAGAGCAAGTGTTCAAATACTTCAAGTGCTTCCAGAAAAACTCGCCTGTGCGCATGCAACTGTATTAG
Protein
MTSKVLLSCVVLAVLATTVLAEDSRKLVSFAPEVAKKLKVLIQECLNENGLGEDAIEVIRAGEYREDEPFQNLVYCAYKKFGALDENNRIISQVAAASFPKDIDVVTVIESCGKEDGNTPVEQVFKYFKCFQKNSPVRMQLY
Summary
Uniprot
H9IZE4
C0SQ81
A0A2K8GKL5
A0A2K8GKM7
A0A2K8GKN1
A0A0L7L7A8
+ More
H9IZE5 A0A2K8GKN7 A0A076E7E0 A0A1E1W8T4 M9YYA4 U5KCW9 A0A076ED46 A0A1B3P5J6 Q5MGD0 A0A0M4JRC2 A0A212EL95 H9IZI1 F1CEY0 A0A2H1VJ21 A0A0S3J2L4 A0A2H1V4I0 A0A0E3Y6X8 A0A310SBA3 H9IZE1 L7X1P8 I1VZV8 Q2F5L4 M4T8H7 A0A2A4JVQ6 A0A1L2BLG4 C7G4W1 A0A1Z2RQG6 A0A2H4ZB03 A0A1V1WC16 A0A0K8TUB3 D6WM83 A0A1B3P5J0 A0A1I9HZL2 A0A0G2YIS6 A0A0M4JIP8 A0A194R2F3 A0A1L2BL98 A0A1I8NGU9 W8BUI1 A0A1D6WII5 W8W3U3 A0A182IYF5 A0A0G2YGV6 A0A2K8GKM6 Q25000 A0A1V1WC13 A0A1J0KKF9 A0A0X8U7T9 A0A0X8VWL7 A0A2P9JZF8 A0A0H3W574 I1VJ75 A0A212EXE2 D4AHN3 A0A0P0UW39 I4DJ69 G9I574 Q8WRV9 A0A2K8GKP3 A0A345BEQ5 Q8WRV8 A0A248Q495 Q8WRW0 A0A1J0KKY1 A0A2H4ZB33 A0A0S3J2I5 A0A2A4ITG4 A0A3F2Z3G8 C7EX19 A0A3L9LY87 D2D0C5 A0A2D1LVP0 A0A1W6L1B0 M9YW76 I1VZV9 A0A2Z4SW61 D2SNV4 A0A109PNL0 L7X1X8
H9IZE5 A0A2K8GKN7 A0A076E7E0 A0A1E1W8T4 M9YYA4 U5KCW9 A0A076ED46 A0A1B3P5J6 Q5MGD0 A0A0M4JRC2 A0A212EL95 H9IZI1 F1CEY0 A0A2H1VJ21 A0A0S3J2L4 A0A2H1V4I0 A0A0E3Y6X8 A0A310SBA3 H9IZE1 L7X1P8 I1VZV8 Q2F5L4 M4T8H7 A0A2A4JVQ6 A0A1L2BLG4 C7G4W1 A0A1Z2RQG6 A0A2H4ZB03 A0A1V1WC16 A0A0K8TUB3 D6WM83 A0A1B3P5J0 A0A1I9HZL2 A0A0G2YIS6 A0A0M4JIP8 A0A194R2F3 A0A1L2BL98 A0A1I8NGU9 W8BUI1 A0A1D6WII5 W8W3U3 A0A182IYF5 A0A0G2YGV6 A0A2K8GKM6 Q25000 A0A1V1WC13 A0A1J0KKF9 A0A0X8U7T9 A0A0X8VWL7 A0A2P9JZF8 A0A0H3W574 I1VJ75 A0A212EXE2 D4AHN3 A0A0P0UW39 I4DJ69 G9I574 Q8WRV9 A0A2K8GKP3 A0A345BEQ5 Q8WRV8 A0A248Q495 Q8WRW0 A0A1J0KKY1 A0A2H4ZB33 A0A0S3J2I5 A0A2A4ITG4 A0A3F2Z3G8 C7EX19 A0A3L9LY87 D2D0C5 A0A2D1LVP0 A0A1W6L1B0 M9YW76 I1VZV9 A0A2Z4SW61 D2SNV4 A0A109PNL0 L7X1X8
Pubmed
EMBL
BABH01014229
AB353891
BAH36760.1
KX585285
ARO70178.1
KX585290
+ More
ARO70183.1 KX585287 ARO70180.1 JTDY01002515 KOB71255.1 BABH01014228 KX585289 ARO70182.1 KF487604 AII01002.1 GDQN01007773 JAT83281.1 KC492498 KC763854 AGK24577.1 AGM38611.1 JX863693 AGR39568.1 KF487582 AII00980.1 KX655922 AOG12871.1 AY829856 AAV91470.1 KT192032 KT261659 ALD65885.1 ALJ30200.1 AGBW02014105 OWR42255.1 BABH01014234 HQ234489 ADY17884.1 ODYU01002843 SOQ40827.1 KT381489 ALR72495.1 ODYU01000620 SOQ35719.1 KM251649 AKC58530.1 GGOB01000085 MCH29481.1 BABH01014235 KC111817 AGC92792.1 JQ753075 AFI57166.1 DQ311409 ABD36353.1 JX962792 AGH70105.1 NWSH01000549 PCG75714.1 KT327225 ALS03868.1 AB518886 BAI22690.1 KX272639 ASA46120.1 MF975370 AUF72946.1 GENK01000029 JAV45884.1 GCVX01000158 JAI18072.1 KQ971343 EFA04593.1 KX655915 AOG12864.1 KF977559 AIX97021.1 KR733547 AKI84359.1 KT192033 KT261668 ALD65886.1 ALJ30209.1 KQ460870 KPJ11699.1 KT327221 ALS03864.1 GAMC01003683 JAC02873.1 KJ082104 AIY61044.1 HG764551 CDJ79887.1 KR733564 AKI84376.1 KX585292 ARO70185.1 L41640 AAA85090.1 GENK01000001 JAV45912.1 KX290639 APC94210.1 KJ186818 AIX97130.1 KJ186822 AIX97134.1 KY826472 AVI04900.1 KP736110 AKK25132.1 BT127821 JQ855696 AEE62783.1 AFI45059.1 AGBW02011795 OWR46117.1 AB521990 BAI82443.1 LC042109 BAS69443.1 AK401337 BAM17959.1 JN571545 AEX07280.1 AF393500 AAL60425.1 KX585312 ARO70205.1 MG788211 AXF48729.1 AF393501 AAL60426.1 KY463449 MH937239 ASM41501.1 AYN70654.1 AF393499 AAL60424.1 KX298775 APC94292.1 MF975394 AUF72970.1 KT381502 ALR72508.1 NWSH01008731 PCG62433.1 GQ301889 ACT75666.1 KB466357 RLZ02248.1 FJ233073 ACO83215.1 KY198524 ATO58974.1 KX609453 KB465664 ARN17860.1 RLZ02146.1 KC492499 AGK24578.1 JQ753076 KZ150094 AFI57167.1 PZC73630.1 MF624623 AWY62809.1 EZ407238 ACX53795.1 KT250756 ALZ41700.1 KC111819 AGC92794.1
ARO70183.1 KX585287 ARO70180.1 JTDY01002515 KOB71255.1 BABH01014228 KX585289 ARO70182.1 KF487604 AII01002.1 GDQN01007773 JAT83281.1 KC492498 KC763854 AGK24577.1 AGM38611.1 JX863693 AGR39568.1 KF487582 AII00980.1 KX655922 AOG12871.1 AY829856 AAV91470.1 KT192032 KT261659 ALD65885.1 ALJ30200.1 AGBW02014105 OWR42255.1 BABH01014234 HQ234489 ADY17884.1 ODYU01002843 SOQ40827.1 KT381489 ALR72495.1 ODYU01000620 SOQ35719.1 KM251649 AKC58530.1 GGOB01000085 MCH29481.1 BABH01014235 KC111817 AGC92792.1 JQ753075 AFI57166.1 DQ311409 ABD36353.1 JX962792 AGH70105.1 NWSH01000549 PCG75714.1 KT327225 ALS03868.1 AB518886 BAI22690.1 KX272639 ASA46120.1 MF975370 AUF72946.1 GENK01000029 JAV45884.1 GCVX01000158 JAI18072.1 KQ971343 EFA04593.1 KX655915 AOG12864.1 KF977559 AIX97021.1 KR733547 AKI84359.1 KT192033 KT261668 ALD65886.1 ALJ30209.1 KQ460870 KPJ11699.1 KT327221 ALS03864.1 GAMC01003683 JAC02873.1 KJ082104 AIY61044.1 HG764551 CDJ79887.1 KR733564 AKI84376.1 KX585292 ARO70185.1 L41640 AAA85090.1 GENK01000001 JAV45912.1 KX290639 APC94210.1 KJ186818 AIX97130.1 KJ186822 AIX97134.1 KY826472 AVI04900.1 KP736110 AKK25132.1 BT127821 JQ855696 AEE62783.1 AFI45059.1 AGBW02011795 OWR46117.1 AB521990 BAI82443.1 LC042109 BAS69443.1 AK401337 BAM17959.1 JN571545 AEX07280.1 AF393500 AAL60425.1 KX585312 ARO70205.1 MG788211 AXF48729.1 AF393501 AAL60426.1 KY463449 MH937239 ASM41501.1 AYN70654.1 AF393499 AAL60424.1 KX298775 APC94292.1 MF975394 AUF72970.1 KT381502 ALR72508.1 NWSH01008731 PCG62433.1 GQ301889 ACT75666.1 KB466357 RLZ02248.1 FJ233073 ACO83215.1 KY198524 ATO58974.1 KX609453 KB465664 ARN17860.1 RLZ02146.1 KC492499 AGK24578.1 JQ753076 KZ150094 AFI57167.1 PZC73630.1 MF624623 AWY62809.1 EZ407238 ACX53795.1 KT250756 ALZ41700.1 KC111819 AGC92794.1
Proteomes
Pfam
PF01395 PBP_GOBP
SUPFAM
SSF47565
SSF47565
Gene 3D
ProteinModelPortal
H9IZE4
C0SQ81
A0A2K8GKL5
A0A2K8GKM7
A0A2K8GKN1
A0A0L7L7A8
+ More
H9IZE5 A0A2K8GKN7 A0A076E7E0 A0A1E1W8T4 M9YYA4 U5KCW9 A0A076ED46 A0A1B3P5J6 Q5MGD0 A0A0M4JRC2 A0A212EL95 H9IZI1 F1CEY0 A0A2H1VJ21 A0A0S3J2L4 A0A2H1V4I0 A0A0E3Y6X8 A0A310SBA3 H9IZE1 L7X1P8 I1VZV8 Q2F5L4 M4T8H7 A0A2A4JVQ6 A0A1L2BLG4 C7G4W1 A0A1Z2RQG6 A0A2H4ZB03 A0A1V1WC16 A0A0K8TUB3 D6WM83 A0A1B3P5J0 A0A1I9HZL2 A0A0G2YIS6 A0A0M4JIP8 A0A194R2F3 A0A1L2BL98 A0A1I8NGU9 W8BUI1 A0A1D6WII5 W8W3U3 A0A182IYF5 A0A0G2YGV6 A0A2K8GKM6 Q25000 A0A1V1WC13 A0A1J0KKF9 A0A0X8U7T9 A0A0X8VWL7 A0A2P9JZF8 A0A0H3W574 I1VJ75 A0A212EXE2 D4AHN3 A0A0P0UW39 I4DJ69 G9I574 Q8WRV9 A0A2K8GKP3 A0A345BEQ5 Q8WRV8 A0A248Q495 Q8WRW0 A0A1J0KKY1 A0A2H4ZB33 A0A0S3J2I5 A0A2A4ITG4 A0A3F2Z3G8 C7EX19 A0A3L9LY87 D2D0C5 A0A2D1LVP0 A0A1W6L1B0 M9YW76 I1VZV9 A0A2Z4SW61 D2SNV4 A0A109PNL0 L7X1X8
H9IZE5 A0A2K8GKN7 A0A076E7E0 A0A1E1W8T4 M9YYA4 U5KCW9 A0A076ED46 A0A1B3P5J6 Q5MGD0 A0A0M4JRC2 A0A212EL95 H9IZI1 F1CEY0 A0A2H1VJ21 A0A0S3J2L4 A0A2H1V4I0 A0A0E3Y6X8 A0A310SBA3 H9IZE1 L7X1P8 I1VZV8 Q2F5L4 M4T8H7 A0A2A4JVQ6 A0A1L2BLG4 C7G4W1 A0A1Z2RQG6 A0A2H4ZB03 A0A1V1WC16 A0A0K8TUB3 D6WM83 A0A1B3P5J0 A0A1I9HZL2 A0A0G2YIS6 A0A0M4JIP8 A0A194R2F3 A0A1L2BL98 A0A1I8NGU9 W8BUI1 A0A1D6WII5 W8W3U3 A0A182IYF5 A0A0G2YGV6 A0A2K8GKM6 Q25000 A0A1V1WC13 A0A1J0KKF9 A0A0X8U7T9 A0A0X8VWL7 A0A2P9JZF8 A0A0H3W574 I1VJ75 A0A212EXE2 D4AHN3 A0A0P0UW39 I4DJ69 G9I574 Q8WRV9 A0A2K8GKP3 A0A345BEQ5 Q8WRV8 A0A248Q495 Q8WRW0 A0A1J0KKY1 A0A2H4ZB33 A0A0S3J2I5 A0A2A4ITG4 A0A3F2Z3G8 C7EX19 A0A3L9LY87 D2D0C5 A0A2D1LVP0 A0A1W6L1B0 M9YW76 I1VZV9 A0A2Z4SW61 D2SNV4 A0A109PNL0 L7X1X8
PDB
5EL2
E-value=7.27884e-05,
Score=103
Ontologies
GO
Topology
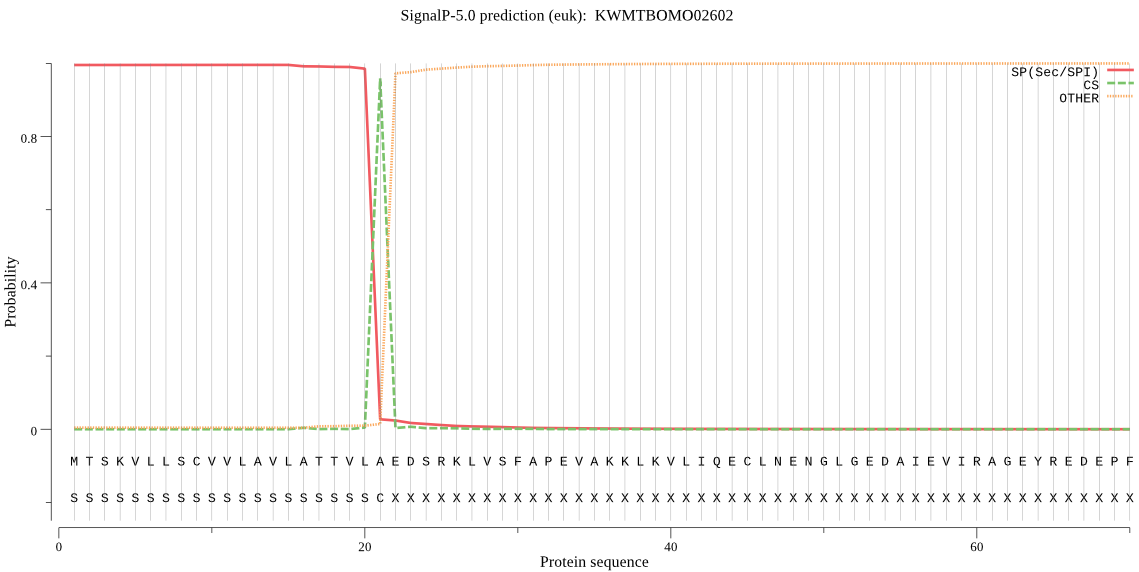
SignalP
Position: 1 - 21,
Likelihood: 0.995326
Length:
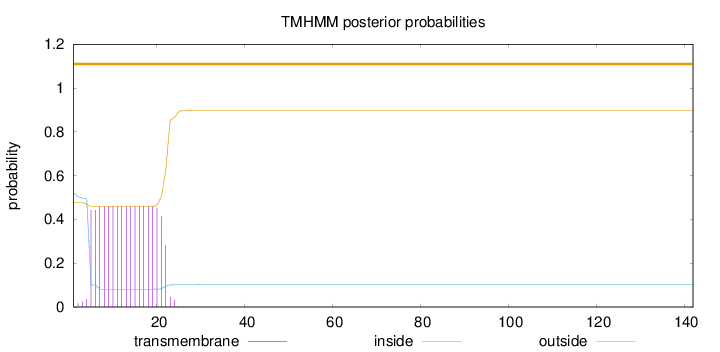
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.19634999999999
Exp number, first 60 AAs:
8.19615
Total prob of N-in:
0.52179
outside
1 - 142
Population Genetic Test Statistics
Pi
195.521675
Theta
149.671509
Tajima's D
1.215597
CLR
0
CSRT
0.71986400679966
Interpretation
Uncertain
