Gene
KWMTBOMO02600
Pre Gene Modal
BGIBMGA002666
Annotation
PREDICTED:_general_odorant-binding_protein_56d-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.084 Extracellular Reliability : 1.517 Nuclear Reliability : 1.42
Sequence
CDS
ATGATAATTATTTTATCATATTCACAGAACGTCCACCTTGCTGAAACTCAAAAAGAAAAAGCTAAGCAGTACACCTCAGAATGTGTTAGAGAATCTGGCGTGAGCACCGAAGCGATAAATGCAGCGAAGATTGGAAAGTACTCCAAAGATAAAGCTTTTAAGAACTTCGTGCTTTGCTTTTTCAAAAAATCCGCAATCTTCAACTCAGATGGTACATTGAACATGGATGTTGCGCTAGCAAAACTTCCTCCTGGTGTTAATAAATCTGAAGCCCAAAGCGTATTAAAACAGTGCAAGAATAAGACCGGCCAAGGCGCAGCCGATAAAGCCTTCGAGATCTTCCGATGTTACTACAAAGGGACCAAGACACATATTTTATTTTAA
Protein
MIIILSYSQNVHLAETQKEKAKQYTSECVRESGVSTEAINAAKIGKYSKDKAFKNFVLCFFKKSAIFNSDGTLNMDVALAKLPPGVNKSEAQSVLKQCKNKTGQGAADKAFEIFRCYYKGTKTHILF
Summary
Uniprot
H9IZI1
C7G4W1
H9IZE1
Q2F5L4
H9IZE2
A0A076E927
+ More
A0A2H4FXY6 Q8WRV9 A0A1B3P5L8 A0A1B4ZBI7 A0A0U3AN25 A0A2K8GKL6 A0A076E7E2 A0A212EL95 A0A194R2F3 A0A2H1V4I0 A0A2H1VJ21 A0A0K8TVG9 A0A0K8TUX5 A0A345BER7 A0A1L2BL94 I4DJ69 E3UPD2 A0A386H995 U5KCW9 F1CEY0 A0A1V1WC13 A0A0M4JRC2 D2SNV4 A0A2A4ITG4 G9I574 I1VZV9 A0A0L7L7D4 L7X1X8 A0A0X8U7T9 F1DI86 E9J206 A0A1C8JZL4 A0A0A7HIY9 A0A142K318 A0A0B4KZC6 A0A3L9LY87 A0A0M4JIP8 M4VTK9 A0A0G2YGV6 A0A1Y0KDK0 A0A0K8TUB3 A0A0X8VWL7 A0A0G2YDB7 A0A0G2YKK7 I1VZV8 A0A0G2YDA5 A0A2P9JZE5 L7X1P8 M3TYZ1 G4Y3X2 A0A1J0KKD4 Q25000 A0A1S5VFG9 M9YW76 A0A1Q1N694 J3JVJ5 A0A232FFM5 A0A0H3W5L4 A0A2A4JVQ6 R9PSN6 G8B1M3 W5JM47 A0A1L2BL98 A0A1V1WC16 A0A1D6WII5 A0A0X9EZ19 A0A0H3W574 A0A182FVB3 A0A1W6L179 I1VJ75 A0A165EY30 A0A1S5VFH1 A0A2P9JZF4 M4T8H7 Q8WRV8 A0A1B1FKI3 A0A1B3P5J0 A0A1C8K2S9 Q7Z1W8 A0A1L8EDQ8 A0A1L8EDJ0 A0A0N9K2E9 A0A1L8EFE6 A0A1L8EFS8 A0A1B0D4J6 U4UHI4 A0A1L8EFR8 A0A194RJX2 I4DJV7 A0A0B4KZD0 Q6F453 B4KRK9 A0A173ACY0
A0A2H4FXY6 Q8WRV9 A0A1B3P5L8 A0A1B4ZBI7 A0A0U3AN25 A0A2K8GKL6 A0A076E7E2 A0A212EL95 A0A194R2F3 A0A2H1V4I0 A0A2H1VJ21 A0A0K8TVG9 A0A0K8TUX5 A0A345BER7 A0A1L2BL94 I4DJ69 E3UPD2 A0A386H995 U5KCW9 F1CEY0 A0A1V1WC13 A0A0M4JRC2 D2SNV4 A0A2A4ITG4 G9I574 I1VZV9 A0A0L7L7D4 L7X1X8 A0A0X8U7T9 F1DI86 E9J206 A0A1C8JZL4 A0A0A7HIY9 A0A142K318 A0A0B4KZC6 A0A3L9LY87 A0A0M4JIP8 M4VTK9 A0A0G2YGV6 A0A1Y0KDK0 A0A0K8TUB3 A0A0X8VWL7 A0A0G2YDB7 A0A0G2YKK7 I1VZV8 A0A0G2YDA5 A0A2P9JZE5 L7X1P8 M3TYZ1 G4Y3X2 A0A1J0KKD4 Q25000 A0A1S5VFG9 M9YW76 A0A1Q1N694 J3JVJ5 A0A232FFM5 A0A0H3W5L4 A0A2A4JVQ6 R9PSN6 G8B1M3 W5JM47 A0A1L2BL98 A0A1V1WC16 A0A1D6WII5 A0A0X9EZ19 A0A0H3W574 A0A182FVB3 A0A1W6L179 I1VJ75 A0A165EY30 A0A1S5VFH1 A0A2P9JZF4 M4T8H7 Q8WRV8 A0A1B1FKI3 A0A1B3P5J0 A0A1C8K2S9 Q7Z1W8 A0A1L8EDQ8 A0A1L8EDJ0 A0A0N9K2E9 A0A1L8EFE6 A0A1L8EFS8 A0A1B0D4J6 U4UHI4 A0A1L8EFR8 A0A194RJX2 I4DJV7 A0A0B4KZD0 Q6F453 B4KRK9 A0A173ACY0
Pubmed
19121390
24998398
27538507
26657286
28736530
22118469
+ More
26354079 26017144 29727827 22651552 24053512 26445454 26346731 20074338 28756777 26227816 21305009 21282665 27652115 25461610 25856077 27012867 23512006 25099623 23517120 27836740 28187311 27545810 22516182 23537049 25938508 28648823 20075255 20920257 23761445 23499610 15303324 26164593 17994087
26354079 26017144 29727827 22651552 24053512 26445454 26346731 20074338 28756777 26227816 21305009 21282665 27652115 25461610 25856077 27012867 23512006 25099623 23517120 27836740 28187311 27545810 22516182 23537049 25938508 28648823 20075255 20920257 23761445 23499610 15303324 26164593 17994087
EMBL
BABH01014234
AB518886
BAI22690.1
BABH01014235
DQ311409
ABD36353.1
+ More
BABH01014233 KF487581 AII00979.1 KY130464 APG32532.1 AF393500 AAL60425.1 KX655930 AOG12879.1 LC027690 BAV56793.1 KP975129 ALT31648.1 KX585283 ARO70176.1 KF487609 AII01007.1 AGBW02014105 OWR42255.1 KQ460870 KPJ11699.1 ODYU01000620 SOQ35719.1 ODYU01002843 SOQ40827.1 GCVX01000160 JAI18070.1 GCVX01000159 JAI18071.1 MG788223 AXF48741.1 KT327213 ALS03856.1 AK401337 BAM17959.1 HM536616 ADO95155.1 MG546557 AYD42180.1 JX863693 AGR39568.1 HQ234489 ADY17884.1 GENK01000001 JAV45912.1 KT192032 KT261659 ALD65885.1 ALJ30200.1 EZ407238 ACX53795.1 NWSH01008731 PCG62433.1 JN571545 AEX07280.1 JQ753076 KZ150094 AFI57167.1 PZC73630.1 JTDY01002515 KOB71254.1 KC111819 AGC92794.1 KJ186818 AIX97130.1 HQ853355 ADX94403.1 GL767709 EFZ13147.1 KR780577 ANE37551.1 KM258401 KM251650 AIZ03625.1 AKC58531.1 KT861417 AMR98353.1 KF383275 AHE13793.1 KB466357 RLZ02248.1 KT192033 KT261668 ALD65886.1 ALJ30209.1 KC464551 AGI05202.1 KR733564 AKI84376.1 KX660672 ARU83754.1 GCVX01000158 JAI18072.1 KJ186822 AIX97134.1 KR733568 AKI84380.1 KR733556 AKI84368.1 JQ753075 AFI57166.1 KR733558 AKI84370.1 KY826458 AVI04886.1 KC111817 AGC92792.1 GACR01000063 JAA74398.1 JF937664 AEP27187.1 KX290616 APC94187.1 L41640 AAA85090.1 KY445447 AQN78382.1 KC492499 AGK24578.1 KX078771 AQM49875.1 APGK01035161 BT127263 KP736119 KB740923 KB632326 AEE62225.1 AKK25138.1 ENN78201.1 ERL92485.1 NNAY01000282 OXU29502.1 KP736125 AKK25143.1 NWSH01000549 PCG75714.1 GABX01000023 JAA74496.1 HE578203 CCD17787.1 ADMH02001024 ETN64378.1 KT327221 ALS03864.1 GENK01000029 JAV45884.1 KJ082104 AIY61044.1 KP453838 ALM64969.1 KP736110 AKK25132.1 KX609458 ARN17865.1 BT127821 JQ855696 AEE62783.1 AFI45059.1 KT983813 AMY16434.1 KY445446 AQN78381.1 KY826468 AVI04896.1 JX962792 AGH70105.1 AF393501 AAL60426.1 KT268307 ANQ46501.1 KX655915 AOG12864.1 KR780581 ANE37555.1 AY182011 AAO64978.1 GFDG01001987 JAV16812.1 GFDG01002018 JAV16781.1 KP963691 ALG36139.1 GFDG01001388 JAV17411.1 GFDG01001376 JAV17423.1 AJVK01003282 ERL92487.1 GFDG01001360 JAV17439.1 KQ460118 KPJ17734.1 AK401575 BAM18197.1 KF383278 AHE13796.1 AB180437 AB189031 BAD26681.1 BAD52263.1 CH933808 EDW10435.1 KT156682 ANG08535.1
BABH01014233 KF487581 AII00979.1 KY130464 APG32532.1 AF393500 AAL60425.1 KX655930 AOG12879.1 LC027690 BAV56793.1 KP975129 ALT31648.1 KX585283 ARO70176.1 KF487609 AII01007.1 AGBW02014105 OWR42255.1 KQ460870 KPJ11699.1 ODYU01000620 SOQ35719.1 ODYU01002843 SOQ40827.1 GCVX01000160 JAI18070.1 GCVX01000159 JAI18071.1 MG788223 AXF48741.1 KT327213 ALS03856.1 AK401337 BAM17959.1 HM536616 ADO95155.1 MG546557 AYD42180.1 JX863693 AGR39568.1 HQ234489 ADY17884.1 GENK01000001 JAV45912.1 KT192032 KT261659 ALD65885.1 ALJ30200.1 EZ407238 ACX53795.1 NWSH01008731 PCG62433.1 JN571545 AEX07280.1 JQ753076 KZ150094 AFI57167.1 PZC73630.1 JTDY01002515 KOB71254.1 KC111819 AGC92794.1 KJ186818 AIX97130.1 HQ853355 ADX94403.1 GL767709 EFZ13147.1 KR780577 ANE37551.1 KM258401 KM251650 AIZ03625.1 AKC58531.1 KT861417 AMR98353.1 KF383275 AHE13793.1 KB466357 RLZ02248.1 KT192033 KT261668 ALD65886.1 ALJ30209.1 KC464551 AGI05202.1 KR733564 AKI84376.1 KX660672 ARU83754.1 GCVX01000158 JAI18072.1 KJ186822 AIX97134.1 KR733568 AKI84380.1 KR733556 AKI84368.1 JQ753075 AFI57166.1 KR733558 AKI84370.1 KY826458 AVI04886.1 KC111817 AGC92792.1 GACR01000063 JAA74398.1 JF937664 AEP27187.1 KX290616 APC94187.1 L41640 AAA85090.1 KY445447 AQN78382.1 KC492499 AGK24578.1 KX078771 AQM49875.1 APGK01035161 BT127263 KP736119 KB740923 KB632326 AEE62225.1 AKK25138.1 ENN78201.1 ERL92485.1 NNAY01000282 OXU29502.1 KP736125 AKK25143.1 NWSH01000549 PCG75714.1 GABX01000023 JAA74496.1 HE578203 CCD17787.1 ADMH02001024 ETN64378.1 KT327221 ALS03864.1 GENK01000029 JAV45884.1 KJ082104 AIY61044.1 KP453838 ALM64969.1 KP736110 AKK25132.1 KX609458 ARN17865.1 BT127821 JQ855696 AEE62783.1 AFI45059.1 KT983813 AMY16434.1 KY445446 AQN78381.1 KY826468 AVI04896.1 JX962792 AGH70105.1 AF393501 AAL60426.1 KT268307 ANQ46501.1 KX655915 AOG12864.1 KR780581 ANE37555.1 AY182011 AAO64978.1 GFDG01001987 JAV16812.1 GFDG01002018 JAV16781.1 KP963691 ALG36139.1 GFDG01001388 JAV17411.1 GFDG01001376 JAV17423.1 AJVK01003282 ERL92487.1 GFDG01001360 JAV17439.1 KQ460118 KPJ17734.1 AK401575 BAM18197.1 KF383278 AHE13796.1 AB180437 AB189031 BAD26681.1 BAD52263.1 CH933808 EDW10435.1 KT156682 ANG08535.1
Proteomes
Pfam
PF01395 PBP_GOBP
SUPFAM
SSF47565
SSF47565
Gene 3D
ProteinModelPortal
H9IZI1
C7G4W1
H9IZE1
Q2F5L4
H9IZE2
A0A076E927
+ More
A0A2H4FXY6 Q8WRV9 A0A1B3P5L8 A0A1B4ZBI7 A0A0U3AN25 A0A2K8GKL6 A0A076E7E2 A0A212EL95 A0A194R2F3 A0A2H1V4I0 A0A2H1VJ21 A0A0K8TVG9 A0A0K8TUX5 A0A345BER7 A0A1L2BL94 I4DJ69 E3UPD2 A0A386H995 U5KCW9 F1CEY0 A0A1V1WC13 A0A0M4JRC2 D2SNV4 A0A2A4ITG4 G9I574 I1VZV9 A0A0L7L7D4 L7X1X8 A0A0X8U7T9 F1DI86 E9J206 A0A1C8JZL4 A0A0A7HIY9 A0A142K318 A0A0B4KZC6 A0A3L9LY87 A0A0M4JIP8 M4VTK9 A0A0G2YGV6 A0A1Y0KDK0 A0A0K8TUB3 A0A0X8VWL7 A0A0G2YDB7 A0A0G2YKK7 I1VZV8 A0A0G2YDA5 A0A2P9JZE5 L7X1P8 M3TYZ1 G4Y3X2 A0A1J0KKD4 Q25000 A0A1S5VFG9 M9YW76 A0A1Q1N694 J3JVJ5 A0A232FFM5 A0A0H3W5L4 A0A2A4JVQ6 R9PSN6 G8B1M3 W5JM47 A0A1L2BL98 A0A1V1WC16 A0A1D6WII5 A0A0X9EZ19 A0A0H3W574 A0A182FVB3 A0A1W6L179 I1VJ75 A0A165EY30 A0A1S5VFH1 A0A2P9JZF4 M4T8H7 Q8WRV8 A0A1B1FKI3 A0A1B3P5J0 A0A1C8K2S9 Q7Z1W8 A0A1L8EDQ8 A0A1L8EDJ0 A0A0N9K2E9 A0A1L8EFE6 A0A1L8EFS8 A0A1B0D4J6 U4UHI4 A0A1L8EFR8 A0A194RJX2 I4DJV7 A0A0B4KZD0 Q6F453 B4KRK9 A0A173ACY0
A0A2H4FXY6 Q8WRV9 A0A1B3P5L8 A0A1B4ZBI7 A0A0U3AN25 A0A2K8GKL6 A0A076E7E2 A0A212EL95 A0A194R2F3 A0A2H1V4I0 A0A2H1VJ21 A0A0K8TVG9 A0A0K8TUX5 A0A345BER7 A0A1L2BL94 I4DJ69 E3UPD2 A0A386H995 U5KCW9 F1CEY0 A0A1V1WC13 A0A0M4JRC2 D2SNV4 A0A2A4ITG4 G9I574 I1VZV9 A0A0L7L7D4 L7X1X8 A0A0X8U7T9 F1DI86 E9J206 A0A1C8JZL4 A0A0A7HIY9 A0A142K318 A0A0B4KZC6 A0A3L9LY87 A0A0M4JIP8 M4VTK9 A0A0G2YGV6 A0A1Y0KDK0 A0A0K8TUB3 A0A0X8VWL7 A0A0G2YDB7 A0A0G2YKK7 I1VZV8 A0A0G2YDA5 A0A2P9JZE5 L7X1P8 M3TYZ1 G4Y3X2 A0A1J0KKD4 Q25000 A0A1S5VFG9 M9YW76 A0A1Q1N694 J3JVJ5 A0A232FFM5 A0A0H3W5L4 A0A2A4JVQ6 R9PSN6 G8B1M3 W5JM47 A0A1L2BL98 A0A1V1WC16 A0A1D6WII5 A0A0X9EZ19 A0A0H3W574 A0A182FVB3 A0A1W6L179 I1VJ75 A0A165EY30 A0A1S5VFH1 A0A2P9JZF4 M4T8H7 Q8WRV8 A0A1B1FKI3 A0A1B3P5J0 A0A1C8K2S9 Q7Z1W8 A0A1L8EDQ8 A0A1L8EDJ0 A0A0N9K2E9 A0A1L8EFE6 A0A1L8EFS8 A0A1B0D4J6 U4UHI4 A0A1L8EFR8 A0A194RJX2 I4DJV7 A0A0B4KZD0 Q6F453 B4KRK9 A0A173ACY0
PDB
5DIC
E-value=2.81479e-09,
Score=141
Ontologies
GO
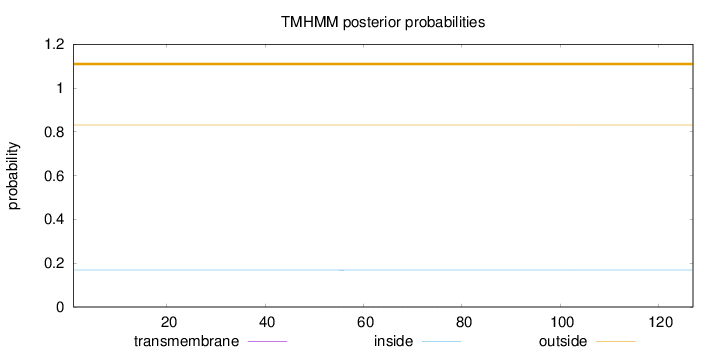
Topology
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01077
Exp number, first 60 AAs:
0.00351
Total prob of N-in:
0.16903
outside
1 - 127
Population Genetic Test Statistics
Pi
310.39426
Theta
179.071315
Tajima's D
2.279999
CLR
0
CSRT
0.92190390480476
Interpretation
Uncertain
