Gene
KWMTBOMO02591
Pre Gene Modal
BGIBMGA002669
Annotation
PREDICTED:_pancreatic_triacylglycerol_lipase-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.681 PlasmaMembrane Reliability : 1.895
Sequence
CDS
ATGTACGGACTGGTTAAGTGTGCGCTGTTACTGTGCGCTGCTACTGCAGCGTACAGTTTCAATTTGGGAGAACGGGATGTTGTATTTCACCTCTTTCATCGGGGAAGCCCTCAGGTCAGTGAACCGTTGTTGCTGTCTGTCAACTCTATTATGACTTCTTCGTTCTCCCTCGCTCGCCGCACCATCTTCACGATACACAATCACGGTGAAACTGTCGCTGGAAACTTCAACGCTTTCGTCATACCCGCTCACCTCGCCGCTGAAGATGTAAACGTAATCGCCGTCGACTGGAGTCCTGGATCGAAATTATACACCGAGGGTCTGGCCAACGCTCGTCAATGCGGTCGATTAATTGCTGAATTCATCAATATCTTGACTACAACCTTCAGCTACGATTCCGAGCTTATTCGCATTGTCGGACTTGGTCTCGGTGGACACATCGCTGGTATCGCAGCTAGAAATTCAAACCAACCTATTCCTCACATTATTGCTCTGGACCCATCTCTCCACGGTTGGACTCACAACCCTGAAATTCTCAATCCTGACGATGCTAACATCGTAGAAGTCCTGCACACAACAGCCGGGCTGATTGGTTATGACTATCCTTTGGGAGATCTCGACTTTTATCCCTCCGGCGGTAGCGGTCAAAGCGGATGCCTGGACGATGCCTGCTCCCATTCTTACGCCTATGTTTTCTACGCCGAATCTATCAGGCAAGCAGTAGATGGAGGTAATGAGTTCGTCGGAACTGCTTGCAATAGCTACGAAGAAGCTCGCAACCTCAGATGCTCTGGACCCAGAGACGCCATTTTCGGTGGAATTGAAGTTAAAGAAGTTGAAATGGGTATATACATATTCTTGACAAACATAAGGCCTCCATTCGCAAGAGATTGA
Protein
MYGLVKCALLLCAATAAYSFNLGERDVVFHLFHRGSPQVSEPLLLSVNSIMTSSFSLARRTIFTIHNHGETVAGNFNAFVIPAHLAAEDVNVIAVDWSPGSKLYTEGLANARQCGRLIAEFINILTTTFSYDSELIRIVGLGLGGHIAGIAARNSNQPIPHIIALDPSLHGWTHNPEILNPDDANIVEVLHTTAGLIGYDYPLGDLDFYPSGGSGQSGCLDDACSHSYAYVFYAESIRQAVDGGNEFVGTACNSYEEARNLRCSGPRDAIFGGIEVKEVEMGIYIFLTNIRPPFARD
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9IZI4
A0A1E1WIQ4
A0A194Q2B8
A0A1E1VZ18
A0A194R386
A0A2A4IVM6
+ More
A0A1E1W7B2 I3QC06 B1NLE6 B6CMF6 A0A212F9I6 F6K6Y6 A0A1E1WV17 A0A0L7LGU1 F5GTF9 A0A2H1VEP8 A0A2W1B5Z2 A0A2A4IWH5 A0A2W1BR39 A0A2W1BPA5 A0A2H1VWT7 A0A2H1VXQ6 B2ZGH7 A0A2A4JT49 A0A1E1W7R1 A0A2A4K7S9 A0A2H1VLE7 A0A2H1VGG1 A0A2H1VU53 A0A2H1W3F9 A0A2A4K841 A0A1E1WHW3 A0A2A4JTH4 V5G104 A0A2W1BTR8 B2ZGH6 A0A2A4JZX6 A0A2W1BND5 B6CMF4 A0A2H1VLN0 A0A2W1BNC9 V5GWU5 D6W6G8 A0A0L7LGM8 B1NLE7 A0A2H1W3D1 I3QC08 A0A195DTF1 A0A1W4XDL3 H9JQT9 A0A1W4WB70 A0A1W4WMA5 A0A2A4IX84 A0A1J1HR28 A0A2A4JES0 E2B986 A0A182JHZ0 A0A182QBK2 A0A2H1V940 A0A0J7MVM6 A0A182IYI0 E9ILQ5 A0A084VK35 F4WF75 A0A182L4C6 T1DJK5 A0A182JLE7 A0A194PI56 A0A1Y9GL11 A0A182VMK6 W5JRH3 A0A182UQU0 A0A2H1WAX6 A0A195F908 A0A1W4WIH6 A0A182GEE7 U4UQR1 B0W1X1 N6UHZ9 J3JV82 Q173Q3 B0WAS0 A0A1W4X462 Q9VB89 A0A1W4X452 A0A194PJT7 A0A182YJC7 D6W6G9 A0A182TKM6 A0A182K7L4 A0A1B0CJD2 B4M128 Q173Q0 B3P873 A0A182KZN8 B3P870 A0A084VIF1 A0A1S4FFB7
A0A1E1W7B2 I3QC06 B1NLE6 B6CMF6 A0A212F9I6 F6K6Y6 A0A1E1WV17 A0A0L7LGU1 F5GTF9 A0A2H1VEP8 A0A2W1B5Z2 A0A2A4IWH5 A0A2W1BR39 A0A2W1BPA5 A0A2H1VWT7 A0A2H1VXQ6 B2ZGH7 A0A2A4JT49 A0A1E1W7R1 A0A2A4K7S9 A0A2H1VLE7 A0A2H1VGG1 A0A2H1VU53 A0A2H1W3F9 A0A2A4K841 A0A1E1WHW3 A0A2A4JTH4 V5G104 A0A2W1BTR8 B2ZGH6 A0A2A4JZX6 A0A2W1BND5 B6CMF4 A0A2H1VLN0 A0A2W1BNC9 V5GWU5 D6W6G8 A0A0L7LGM8 B1NLE7 A0A2H1W3D1 I3QC08 A0A195DTF1 A0A1W4XDL3 H9JQT9 A0A1W4WB70 A0A1W4WMA5 A0A2A4IX84 A0A1J1HR28 A0A2A4JES0 E2B986 A0A182JHZ0 A0A182QBK2 A0A2H1V940 A0A0J7MVM6 A0A182IYI0 E9ILQ5 A0A084VK35 F4WF75 A0A182L4C6 T1DJK5 A0A182JLE7 A0A194PI56 A0A1Y9GL11 A0A182VMK6 W5JRH3 A0A182UQU0 A0A2H1WAX6 A0A195F908 A0A1W4WIH6 A0A182GEE7 U4UQR1 B0W1X1 N6UHZ9 J3JV82 Q173Q3 B0WAS0 A0A1W4X462 Q9VB89 A0A1W4X452 A0A194PJT7 A0A182YJC7 D6W6G9 A0A182TKM6 A0A182K7L4 A0A1B0CJD2 B4M128 Q173Q0 B3P873 A0A182KZN8 B3P870 A0A084VIF1 A0A1S4FFB7
Pubmed
19121390
26354079
18314941
18760362
22118469
26227816
+ More
21910995 28756777 18362917 19820115 20798317 21282665 24438588 21719571 20966253 20920257 23761445 26483478 23537049 22516182 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 17994087
21910995 28756777 18362917 19820115 20798317 21282665 24438588 21719571 20966253 20920257 23761445 26483478 23537049 22516182 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 17994087
EMBL
BABH01014242
BABH01014243
BABH01014244
BABH01014245
BABH01014246
GDQN01004165
+ More
JAT86889.1 KQ459589 KPI97550.1 GDQN01011122 JAT79932.1 KQ460870 KPJ11705.1 NWSH01005705 PCG64027.1 GDQN01008217 JAT82837.1 JF999955 AFI64309.1 EF600061 ABU98626.1 EU325556 ACB54946.1 AGBW02009585 OWR50387.1 HM357823 AEA76289.1 GDQN01000378 JAT90676.1 JTDY01001188 KOB74604.1 JI484232 AEB26283.1 ODYU01002163 SOQ39305.1 KZ150473 PZC70751.1 PCG64029.1 KZ150008 PZC75136.1 PZC75137.1 ODYU01004911 SOQ45273.1 SOQ45272.1 EU660854 ACD37364.1 NWSH01000654 PCG74999.1 GDQN01008012 JAT83042.1 NWSH01000080 PCG79823.1 ODYU01002941 SOQ41074.1 SOQ39304.1 ODYU01004467 SOQ44367.1 ODYU01006078 SOQ47611.1 PCG79822.1 GDQN01004482 JAT86572.1 PCG74998.1 GALX01004776 JAB63690.1 PZC75133.1 EU660853 ACD37363.1 NWSH01000332 PCG77326.1 PZC75135.1 EU325554 ACB54944.1 ODYU01002977 SOQ41154.1 PZC75134.1 GALX01003723 JAB64743.1 KQ971307 EFA11062.1 KOB74607.1 EF600062 ABU98627.1 ODYU01006077 SOQ47610.1 JF999957 AFI64311.1 KQ980419 KYN16118.1 BABH01028718 PCG64028.1 CVRI01000019 CRK90495.1 NWSH01001649 PCG70557.1 GL446473 EFN87734.1 AXCN02001074 ODYU01001301 SOQ37321.1 LBMM01016135 KMQ84535.1 GL764129 EFZ18456.1 ATLV01014098 KE524940 KFB38329.1 GL888115 EGI67126.1 GAMD01001396 JAB00195.1 KQ459603 KPI92733.1 APCN01002368 ADMH02000317 ETN66992.1 ODYU01007448 SOQ50231.1 KQ981727 KYN36866.1 JXUM01057935 KQ561979 KXJ76999.1 KB632399 ERL94833.1 DS231824 EDS27407.1 APGK01024781 KB740562 ENN80261.1 BT127149 AEE62111.1 ERL94835.1 CH477418 EAT41284.1 DS231875 EDS41786.1 AE014297 BT030806 AAF56653.2 ABV82188.1 KQ459602 KPI93582.1 EFA11061.1 AJWK01014495 CH940650 EDW67439.1 EAT41287.1 CH954182 EDV53477.1 EDV53474.1 ATLV01013354 KE524854 KFB37745.1
JAT86889.1 KQ459589 KPI97550.1 GDQN01011122 JAT79932.1 KQ460870 KPJ11705.1 NWSH01005705 PCG64027.1 GDQN01008217 JAT82837.1 JF999955 AFI64309.1 EF600061 ABU98626.1 EU325556 ACB54946.1 AGBW02009585 OWR50387.1 HM357823 AEA76289.1 GDQN01000378 JAT90676.1 JTDY01001188 KOB74604.1 JI484232 AEB26283.1 ODYU01002163 SOQ39305.1 KZ150473 PZC70751.1 PCG64029.1 KZ150008 PZC75136.1 PZC75137.1 ODYU01004911 SOQ45273.1 SOQ45272.1 EU660854 ACD37364.1 NWSH01000654 PCG74999.1 GDQN01008012 JAT83042.1 NWSH01000080 PCG79823.1 ODYU01002941 SOQ41074.1 SOQ39304.1 ODYU01004467 SOQ44367.1 ODYU01006078 SOQ47611.1 PCG79822.1 GDQN01004482 JAT86572.1 PCG74998.1 GALX01004776 JAB63690.1 PZC75133.1 EU660853 ACD37363.1 NWSH01000332 PCG77326.1 PZC75135.1 EU325554 ACB54944.1 ODYU01002977 SOQ41154.1 PZC75134.1 GALX01003723 JAB64743.1 KQ971307 EFA11062.1 KOB74607.1 EF600062 ABU98627.1 ODYU01006077 SOQ47610.1 JF999957 AFI64311.1 KQ980419 KYN16118.1 BABH01028718 PCG64028.1 CVRI01000019 CRK90495.1 NWSH01001649 PCG70557.1 GL446473 EFN87734.1 AXCN02001074 ODYU01001301 SOQ37321.1 LBMM01016135 KMQ84535.1 GL764129 EFZ18456.1 ATLV01014098 KE524940 KFB38329.1 GL888115 EGI67126.1 GAMD01001396 JAB00195.1 KQ459603 KPI92733.1 APCN01002368 ADMH02000317 ETN66992.1 ODYU01007448 SOQ50231.1 KQ981727 KYN36866.1 JXUM01057935 KQ561979 KXJ76999.1 KB632399 ERL94833.1 DS231824 EDS27407.1 APGK01024781 KB740562 ENN80261.1 BT127149 AEE62111.1 ERL94835.1 CH477418 EAT41284.1 DS231875 EDS41786.1 AE014297 BT030806 AAF56653.2 ABV82188.1 KQ459602 KPI93582.1 EFA11061.1 AJWK01014495 CH940650 EDW67439.1 EAT41287.1 CH954182 EDV53477.1 EDV53474.1 ATLV01013354 KE524854 KFB37745.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000007266 UP000078492 UP000192223 UP000183832 UP000008237 UP000075880 UP000075886 UP000036403 UP000030765 UP000007755 UP000075882 UP000075840 UP000075903 UP000000673 UP000078541 UP000069940 UP000249989 UP000030742 UP000002320 UP000019118 UP000008820 UP000000803 UP000076408 UP000075902 UP000075881 UP000092461 UP000008792 UP000008711
UP000007266 UP000078492 UP000192223 UP000183832 UP000008237 UP000075880 UP000075886 UP000036403 UP000030765 UP000007755 UP000075882 UP000075840 UP000075903 UP000000673 UP000078541 UP000069940 UP000249989 UP000030742 UP000002320 UP000019118 UP000008820 UP000000803 UP000076408 UP000075902 UP000075881 UP000092461 UP000008792 UP000008711
Interpro
Gene 3D
ProteinModelPortal
H9IZI4
A0A1E1WIQ4
A0A194Q2B8
A0A1E1VZ18
A0A194R386
A0A2A4IVM6
+ More
A0A1E1W7B2 I3QC06 B1NLE6 B6CMF6 A0A212F9I6 F6K6Y6 A0A1E1WV17 A0A0L7LGU1 F5GTF9 A0A2H1VEP8 A0A2W1B5Z2 A0A2A4IWH5 A0A2W1BR39 A0A2W1BPA5 A0A2H1VWT7 A0A2H1VXQ6 B2ZGH7 A0A2A4JT49 A0A1E1W7R1 A0A2A4K7S9 A0A2H1VLE7 A0A2H1VGG1 A0A2H1VU53 A0A2H1W3F9 A0A2A4K841 A0A1E1WHW3 A0A2A4JTH4 V5G104 A0A2W1BTR8 B2ZGH6 A0A2A4JZX6 A0A2W1BND5 B6CMF4 A0A2H1VLN0 A0A2W1BNC9 V5GWU5 D6W6G8 A0A0L7LGM8 B1NLE7 A0A2H1W3D1 I3QC08 A0A195DTF1 A0A1W4XDL3 H9JQT9 A0A1W4WB70 A0A1W4WMA5 A0A2A4IX84 A0A1J1HR28 A0A2A4JES0 E2B986 A0A182JHZ0 A0A182QBK2 A0A2H1V940 A0A0J7MVM6 A0A182IYI0 E9ILQ5 A0A084VK35 F4WF75 A0A182L4C6 T1DJK5 A0A182JLE7 A0A194PI56 A0A1Y9GL11 A0A182VMK6 W5JRH3 A0A182UQU0 A0A2H1WAX6 A0A195F908 A0A1W4WIH6 A0A182GEE7 U4UQR1 B0W1X1 N6UHZ9 J3JV82 Q173Q3 B0WAS0 A0A1W4X462 Q9VB89 A0A1W4X452 A0A194PJT7 A0A182YJC7 D6W6G9 A0A182TKM6 A0A182K7L4 A0A1B0CJD2 B4M128 Q173Q0 B3P873 A0A182KZN8 B3P870 A0A084VIF1 A0A1S4FFB7
A0A1E1W7B2 I3QC06 B1NLE6 B6CMF6 A0A212F9I6 F6K6Y6 A0A1E1WV17 A0A0L7LGU1 F5GTF9 A0A2H1VEP8 A0A2W1B5Z2 A0A2A4IWH5 A0A2W1BR39 A0A2W1BPA5 A0A2H1VWT7 A0A2H1VXQ6 B2ZGH7 A0A2A4JT49 A0A1E1W7R1 A0A2A4K7S9 A0A2H1VLE7 A0A2H1VGG1 A0A2H1VU53 A0A2H1W3F9 A0A2A4K841 A0A1E1WHW3 A0A2A4JTH4 V5G104 A0A2W1BTR8 B2ZGH6 A0A2A4JZX6 A0A2W1BND5 B6CMF4 A0A2H1VLN0 A0A2W1BNC9 V5GWU5 D6W6G8 A0A0L7LGM8 B1NLE7 A0A2H1W3D1 I3QC08 A0A195DTF1 A0A1W4XDL3 H9JQT9 A0A1W4WB70 A0A1W4WMA5 A0A2A4IX84 A0A1J1HR28 A0A2A4JES0 E2B986 A0A182JHZ0 A0A182QBK2 A0A2H1V940 A0A0J7MVM6 A0A182IYI0 E9ILQ5 A0A084VK35 F4WF75 A0A182L4C6 T1DJK5 A0A182JLE7 A0A194PI56 A0A1Y9GL11 A0A182VMK6 W5JRH3 A0A182UQU0 A0A2H1WAX6 A0A195F908 A0A1W4WIH6 A0A182GEE7 U4UQR1 B0W1X1 N6UHZ9 J3JV82 Q173Q3 B0WAS0 A0A1W4X462 Q9VB89 A0A1W4X452 A0A194PJT7 A0A182YJC7 D6W6G9 A0A182TKM6 A0A182K7L4 A0A1B0CJD2 B4M128 Q173Q0 B3P873 A0A182KZN8 B3P870 A0A084VIF1 A0A1S4FFB7
PDB
1ETH
E-value=5.71486e-22,
Score=256
Ontologies
GO
PANTHER
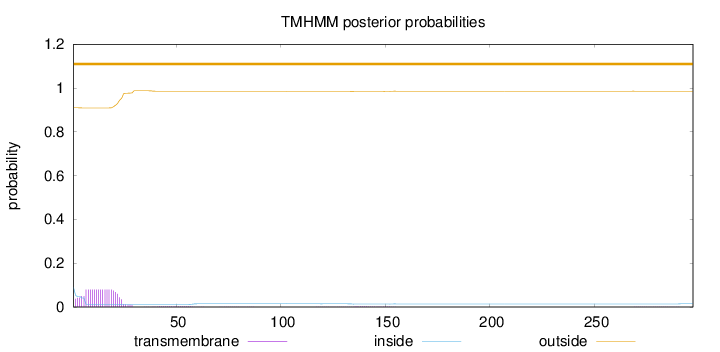
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 19,
Likelihood: 0.929460
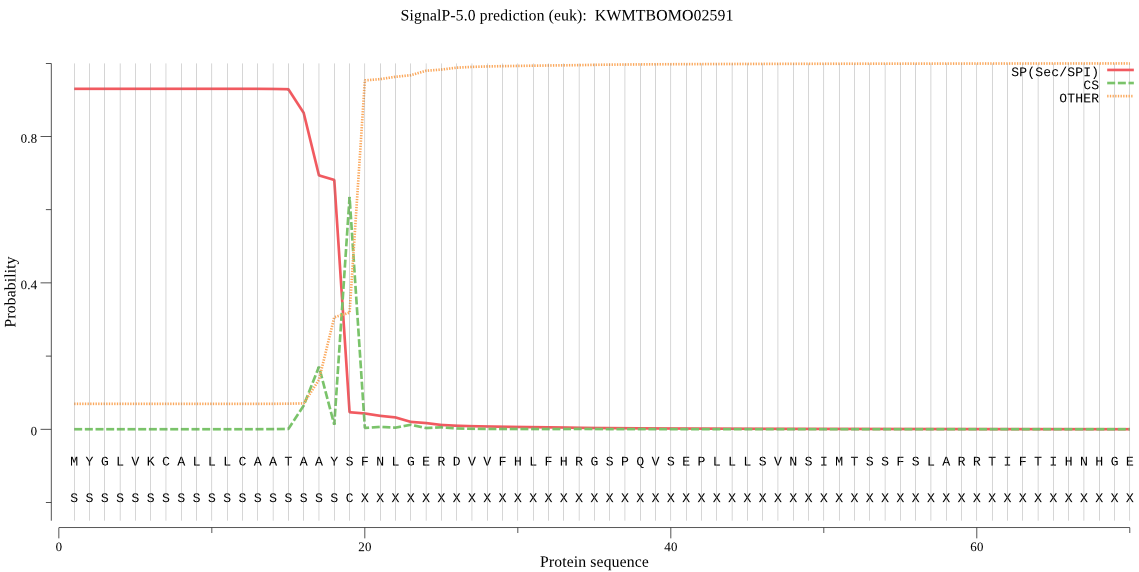
Length:
297
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.71114
Exp number, first 60 AAs:
1.64844
Total prob of N-in:
0.09003
outside
1 - 297
Population Genetic Test Statistics
Pi
294.385923
Theta
164.03738
Tajima's D
2.44236
CLR
0.048303
CSRT
0.941452927353632
Interpretation
Uncertain
