Gene
KWMTBOMO02586
Annotation
PREDICTED:_sodium-coupled_monocarboxylate_transporter_1-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.988
Sequence
CDS
ATGGCGGAAGATAACTTCAAATCGGACGTGGCAGATGTGAGATTTTCCCTCCAACGTTTTACCTATGTAGATTACGCTGTGTTTATTATGATGTTGACAATTTGTGGCTTAATTGGAATATATTTTGGATTCGTGAATAAACAGAAATCAACCCAGGATTACCTTATGGGTGGAAGAAACATGAAATTGGTGCCTGTTTGTTTTTCTTTAGTTGCAAGTTTCATTTCAGGTATATCTCTGCTCGGAACGCCCACAGAACTATATCTGTTCGGGACGGCTTATGTGTTCTCACTGTTCGGCGCCGTAGCAATGTCTCTTATATTAGGATCAACATTTCTACCGGTTTTTCATGAGCTCCAAATAACTTCAGTTTACGAATATTTGGAGCTGCGCTACGATAAACGAGTCAGGATATTTGGTTCAGCTATATTCAGTATATATTTGATGGCTTGGCTGCCTATAGTGATATATGTTCCGGCTTTAGCATTCAATCAAGTGACCGGATTTAACATACACATTATTTCACCGATTATTTGTTCTGTTTGCATTTTCTATACTTCCTTGGGAGGACTCAAAGCGGTTGTATGGACGGATGTGCTTCAAACCATTGTAATGGTATCAGCTATGATTTTAGTTATATTCAAAAGTACAATAATTGTGGGAGGTATCAACGAAGTTTTGAGACGAAACTGGAGTACCGACAGACTCGCGTTTCCATCTTTGACTTTTGATTTAACGGAAAGACACTCAATCTGGTCTGTGTCTATTGGTGCTACATTCTATTGGATTGGGAATATTGCTGTCAATCAATCTATGATGCAGAGGTTCCTCGCTCTCTCAGAGCTAAAGTCTGCTAAACGGGCAATTTGGGGTTTTCTTTTTGGAGTCTTCGTAATAGTTTTTATATGTGGATACAGTGGATTGGTTGCATATGCTAAGTACTATGAGTGCGATCCATTGGATTCAAAATTGGCTCTGGCTAAGGACCAGTTATTGCCGTTGTTGGTGATGGATGTATTTAACGATTGGGAAGGAATGCCCGGAATTTTTATTGCAGGAGTATTCAGCGCTGCTCTAAGTTCGTTATCAACTGGTCTTAATTCTATGGCTGCGGTAGTTTTAGAAGATTTCTGGAAGCCTTTCTGTAAAGTACCGACACCCAAACAATCTCAAATTATTGTTCGTTTAGTCGTCATCATTCTCGGCTGTATTTGTTTTGGTTTAGTGTTCGTAGTTGAAAAACTAGGTTCTGTATTACAATTAACGATGAGTTTATCTTCAGCTTCAATGGGACCTCTTGCAGGAGTGTTTCTGATGGGACTGTTCTTTCCGTTTATAGATTCAACTGATGCACTGTGTGGTGGGATAATTGGATTGCTATCCGCATGGTGGGTAACAGCACAGTCTCAATTTGCGATTGCAAAAGGTCTAATGAAGTATAATGAAAAGATCAGATTCACTAATAACTGTAGTTACACTTTTAAATCAGCAGTCGTTCATGCTACAGAGAGCAGTTCTAGCGAGGTGCCAGCTTTGTACCGTATTTCATATTTATGGTTCACTGCGCTGGGTTGTATTGTAACCGTAACGGTTGCATGTTTAGTTTCGTTAAGGACTTCGAATAAAGTTAAGCGTGACATAAGGACATTTGCTCCTGCTATTAGGAAGTACTTAAACAAACGGGAAGGAAACGATTTCAGCATGGACGAGAAATGTAGATTTAACATTATACCGGAAGCCTTCAATTGA
Protein
MAEDNFKSDVADVRFSLQRFTYVDYAVFIMMLTICGLIGIYFGFVNKQKSTQDYLMGGRNMKLVPVCFSLVASFISGISLLGTPTELYLFGTAYVFSLFGAVAMSLILGSTFLPVFHELQITSVYEYLELRYDKRVRIFGSAIFSIYLMAWLPIVIYVPALAFNQVTGFNIHIISPIICSVCIFYTSLGGLKAVVWTDVLQTIVMVSAMILVIFKSTIIVGGINEVLRRNWSTDRLAFPSLTFDLTERHSIWSVSIGATFYWIGNIAVNQSMMQRFLALSELKSAKRAIWGFLFGVFVIVFICGYSGLVAYAKYYECDPLDSKLALAKDQLLPLLVMDVFNDWEGMPGIFIAGVFSAALSSLSTGLNSMAAVVLEDFWKPFCKVPTPKQSQIIVRLVVIILGCICFGLVFVVEKLGSVLQLTMSLSSASMGPLAGVFLMGLFFPFIDSTDALCGGIIGLLSAWWVTAQSQFAIAKGLMKYNEKIRFTNNCSYTFKSAVVHATESSSSEVPALYRISYLWFTALGCIVTVTVACLVSLRTSNKVKRDIRTFAPAIRKYLNKREGNDFSMDEKCRFNIIPEAFN
Summary
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Uniprot
A0A2A4JGL0
A0A2H1V6Z9
A0A194PX11
A0A194R2G2
H9IZD8
A0A182FS71
+ More
Q7PX28 A0A182L8F2 A0A182QU12 W5J189 A0A182PJ36 A0A182VW47 A0A182HK72 A0A182XP36 A0A0C9R126 Q175Y8 A0A151X8I1 A0A026WWM1 A0A154PEG1 K7J4Z0 A0A182YHT2 A0A0K8V0X3 A0A3B0KJA3 F4W7J6 B5DX93 B4GLN6 B4LXL6 W8BMP0 A0A195ESL3 A0A1B0A1P5 A0A0A1XSV9 A0A182GDR0 A0A1A9VL99 A0A195EL28 A0A1Q3FSJ8 A0A0M8ZZT7 B4NAX6 A0A2P8XTX8 A0A1A9X2N1 A0A0A1WU89 A0A2J7QJP6 Q16LC9 A0A1B0FN19 A0A1I8Q4W1 A0A1Q3FSH6 B4JTK2 A0A2A3EKX1 A0A067QX91 V9IKT4 A0A1B0BA90 A0A2J7QJQ3 W8BJ82 A0A0K8VUV5 A0A084W0F7 A0A232EYM7 A0A1Q3G536 B4K9T9 A0A1A9XW60 A0A195AT73 A0A1W4XEU6 A0A1W4X5A6 A0A2A4JF18 A0A182NQW1 A0A0L0CPB2 A0A1W4WCA6 T1GHV9 B3LXC3 A0A212FLL7 A0A1I8Q8H6 A0A1I8MDU3 A0A182PGU7 A0A1J1HJW9 A0A182VWJ0 A0A0M4F4Q2 A0A1Y9IMI8 A0A182L9J4 A0A182ULS2 A0A0L0C9P3 A0A182WSB4 A0A182HL66 E2A8C5 A0A182SGA4 T1PAX5 A0A1B6DZY1 Q7PXS9 A0A1I8MZG3 A0A034V935 A0A336KMW3 A0A182RSH4 A0A182JVH7 A0A182VI42 A0A182TWQ2 A0A1B0EY77 A0A1L8DET3 A0A1A9ZES2 A0A1B0G0B1 W5J3T7 A0A1A9VCT3 A0A1B6CDJ6 E0VCA6
Q7PX28 A0A182L8F2 A0A182QU12 W5J189 A0A182PJ36 A0A182VW47 A0A182HK72 A0A182XP36 A0A0C9R126 Q175Y8 A0A151X8I1 A0A026WWM1 A0A154PEG1 K7J4Z0 A0A182YHT2 A0A0K8V0X3 A0A3B0KJA3 F4W7J6 B5DX93 B4GLN6 B4LXL6 W8BMP0 A0A195ESL3 A0A1B0A1P5 A0A0A1XSV9 A0A182GDR0 A0A1A9VL99 A0A195EL28 A0A1Q3FSJ8 A0A0M8ZZT7 B4NAX6 A0A2P8XTX8 A0A1A9X2N1 A0A0A1WU89 A0A2J7QJP6 Q16LC9 A0A1B0FN19 A0A1I8Q4W1 A0A1Q3FSH6 B4JTK2 A0A2A3EKX1 A0A067QX91 V9IKT4 A0A1B0BA90 A0A2J7QJQ3 W8BJ82 A0A0K8VUV5 A0A084W0F7 A0A232EYM7 A0A1Q3G536 B4K9T9 A0A1A9XW60 A0A195AT73 A0A1W4XEU6 A0A1W4X5A6 A0A2A4JF18 A0A182NQW1 A0A0L0CPB2 A0A1W4WCA6 T1GHV9 B3LXC3 A0A212FLL7 A0A1I8Q8H6 A0A1I8MDU3 A0A182PGU7 A0A1J1HJW9 A0A182VWJ0 A0A0M4F4Q2 A0A1Y9IMI8 A0A182L9J4 A0A182ULS2 A0A0L0C9P3 A0A182WSB4 A0A182HL66 E2A8C5 A0A182SGA4 T1PAX5 A0A1B6DZY1 Q7PXS9 A0A1I8MZG3 A0A034V935 A0A336KMW3 A0A182RSH4 A0A182JVH7 A0A182VI42 A0A182TWQ2 A0A1B0EY77 A0A1L8DET3 A0A1A9ZES2 A0A1B0G0B1 W5J3T7 A0A1A9VCT3 A0A1B6CDJ6 E0VCA6
Pubmed
EMBL
NWSH01001649
PCG70560.1
ODYU01001007
SOQ36623.1
KQ459589
KPI97553.1
+ More
KQ460870 KPJ11709.1 BABH01014248 BABH01014249 AAAB01008987 EAA00957.5 AXCN02001489 ADMH02002189 ETN57897.1 APCN01000818 GBYB01001680 JAG71447.1 CH477393 EAT41908.1 KQ982409 KYQ56672.1 KK107105 QOIP01000011 EZA59529.1 RLU16728.1 KQ434889 KZC10252.1 GDHF01019732 JAI32582.1 OUUW01000008 SPP83838.1 GL887844 EGI69869.1 CM000070 EDY68625.1 CH479185 EDW38460.1 CH940650 EDW66800.1 GAMC01008302 JAB98253.1 KQ981986 KYN31235.1 GBXI01000171 JAD14121.1 JXUM01056488 JXUM01056489 JXUM01056490 KQ561912 KXJ77184.1 KQ978730 KYN28960.1 GFDL01004500 JAV30545.1 KQ435808 KOX72983.1 CH964232 EDW80940.1 PYGN01001356 PSN35450.1 GBXI01012207 JAD02085.1 NEVH01013550 PNF28807.1 CH477910 EAT35131.1 CCAG010013983 GFDL01004485 JAV30560.1 CH916373 EDV95092.1 KZ288217 PBC32370.1 KK852854 KDR14984.1 JR049336 AEY60929.1 JXJN01010867 PNF28804.1 GAMC01016901 JAB89654.1 GDHF01009661 JAI42653.1 ATLV01019121 KE525262 KFB43701.1 NNAY01001584 OXU23524.1 GFDL01000142 JAV34903.1 CH933806 EDW15587.1 KQ976745 KYM75423.1 PCG70561.1 JRES01000104 KNC34106.1 CAQQ02015090 CH902617 EDV42767.2 AGBW02007743 OWR54626.1 CVRI01000002 CRK86766.1 CP012526 ALC46370.1 JRES01000819 KNC28169.1 APCN01000881 GL437536 EFN70310.1 KA645849 AFP60478.1 GEDC01006078 JAS31220.1 EAA00833.5 GAKP01020702 JAC38250.1 UFQS01000652 UFQT01000652 SSX05789.1 SSX26148.1 AJVK01017786 GFDF01009115 JAV04969.1 CCAG010002737 ADMH02002131 ETN58506.1 GEDC01025774 JAS11524.1 DS235051 EEB11028.1
KQ460870 KPJ11709.1 BABH01014248 BABH01014249 AAAB01008987 EAA00957.5 AXCN02001489 ADMH02002189 ETN57897.1 APCN01000818 GBYB01001680 JAG71447.1 CH477393 EAT41908.1 KQ982409 KYQ56672.1 KK107105 QOIP01000011 EZA59529.1 RLU16728.1 KQ434889 KZC10252.1 GDHF01019732 JAI32582.1 OUUW01000008 SPP83838.1 GL887844 EGI69869.1 CM000070 EDY68625.1 CH479185 EDW38460.1 CH940650 EDW66800.1 GAMC01008302 JAB98253.1 KQ981986 KYN31235.1 GBXI01000171 JAD14121.1 JXUM01056488 JXUM01056489 JXUM01056490 KQ561912 KXJ77184.1 KQ978730 KYN28960.1 GFDL01004500 JAV30545.1 KQ435808 KOX72983.1 CH964232 EDW80940.1 PYGN01001356 PSN35450.1 GBXI01012207 JAD02085.1 NEVH01013550 PNF28807.1 CH477910 EAT35131.1 CCAG010013983 GFDL01004485 JAV30560.1 CH916373 EDV95092.1 KZ288217 PBC32370.1 KK852854 KDR14984.1 JR049336 AEY60929.1 JXJN01010867 PNF28804.1 GAMC01016901 JAB89654.1 GDHF01009661 JAI42653.1 ATLV01019121 KE525262 KFB43701.1 NNAY01001584 OXU23524.1 GFDL01000142 JAV34903.1 CH933806 EDW15587.1 KQ976745 KYM75423.1 PCG70561.1 JRES01000104 KNC34106.1 CAQQ02015090 CH902617 EDV42767.2 AGBW02007743 OWR54626.1 CVRI01000002 CRK86766.1 CP012526 ALC46370.1 JRES01000819 KNC28169.1 APCN01000881 GL437536 EFN70310.1 KA645849 AFP60478.1 GEDC01006078 JAS31220.1 EAA00833.5 GAKP01020702 JAC38250.1 UFQS01000652 UFQT01000652 SSX05789.1 SSX26148.1 AJVK01017786 GFDF01009115 JAV04969.1 CCAG010002737 ADMH02002131 ETN58506.1 GEDC01025774 JAS11524.1 DS235051 EEB11028.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000005204
UP000069272
UP000007062
+ More
UP000075882 UP000075886 UP000000673 UP000075885 UP000075920 UP000075840 UP000076407 UP000008820 UP000075809 UP000053097 UP000279307 UP000076502 UP000002358 UP000076408 UP000268350 UP000007755 UP000001819 UP000008744 UP000008792 UP000078541 UP000092445 UP000069940 UP000249989 UP000078200 UP000078492 UP000053105 UP000007798 UP000245037 UP000091820 UP000235965 UP000092444 UP000095300 UP000001070 UP000242457 UP000027135 UP000092460 UP000030765 UP000215335 UP000009192 UP000092443 UP000078540 UP000192223 UP000075884 UP000037069 UP000192221 UP000015102 UP000007801 UP000007151 UP000095301 UP000183832 UP000092553 UP000075902 UP000075903 UP000000311 UP000075901 UP000075900 UP000075881 UP000092462 UP000009046
UP000075882 UP000075886 UP000000673 UP000075885 UP000075920 UP000075840 UP000076407 UP000008820 UP000075809 UP000053097 UP000279307 UP000076502 UP000002358 UP000076408 UP000268350 UP000007755 UP000001819 UP000008744 UP000008792 UP000078541 UP000092445 UP000069940 UP000249989 UP000078200 UP000078492 UP000053105 UP000007798 UP000245037 UP000091820 UP000235965 UP000092444 UP000095300 UP000001070 UP000242457 UP000027135 UP000092460 UP000030765 UP000215335 UP000009192 UP000092443 UP000078540 UP000192223 UP000075884 UP000037069 UP000192221 UP000015102 UP000007801 UP000007151 UP000095301 UP000183832 UP000092553 UP000075902 UP000075903 UP000000311 UP000075901 UP000075900 UP000075881 UP000092462 UP000009046
PRIDE
Pfam
PF00474 SSF
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JGL0
A0A2H1V6Z9
A0A194PX11
A0A194R2G2
H9IZD8
A0A182FS71
+ More
Q7PX28 A0A182L8F2 A0A182QU12 W5J189 A0A182PJ36 A0A182VW47 A0A182HK72 A0A182XP36 A0A0C9R126 Q175Y8 A0A151X8I1 A0A026WWM1 A0A154PEG1 K7J4Z0 A0A182YHT2 A0A0K8V0X3 A0A3B0KJA3 F4W7J6 B5DX93 B4GLN6 B4LXL6 W8BMP0 A0A195ESL3 A0A1B0A1P5 A0A0A1XSV9 A0A182GDR0 A0A1A9VL99 A0A195EL28 A0A1Q3FSJ8 A0A0M8ZZT7 B4NAX6 A0A2P8XTX8 A0A1A9X2N1 A0A0A1WU89 A0A2J7QJP6 Q16LC9 A0A1B0FN19 A0A1I8Q4W1 A0A1Q3FSH6 B4JTK2 A0A2A3EKX1 A0A067QX91 V9IKT4 A0A1B0BA90 A0A2J7QJQ3 W8BJ82 A0A0K8VUV5 A0A084W0F7 A0A232EYM7 A0A1Q3G536 B4K9T9 A0A1A9XW60 A0A195AT73 A0A1W4XEU6 A0A1W4X5A6 A0A2A4JF18 A0A182NQW1 A0A0L0CPB2 A0A1W4WCA6 T1GHV9 B3LXC3 A0A212FLL7 A0A1I8Q8H6 A0A1I8MDU3 A0A182PGU7 A0A1J1HJW9 A0A182VWJ0 A0A0M4F4Q2 A0A1Y9IMI8 A0A182L9J4 A0A182ULS2 A0A0L0C9P3 A0A182WSB4 A0A182HL66 E2A8C5 A0A182SGA4 T1PAX5 A0A1B6DZY1 Q7PXS9 A0A1I8MZG3 A0A034V935 A0A336KMW3 A0A182RSH4 A0A182JVH7 A0A182VI42 A0A182TWQ2 A0A1B0EY77 A0A1L8DET3 A0A1A9ZES2 A0A1B0G0B1 W5J3T7 A0A1A9VCT3 A0A1B6CDJ6 E0VCA6
Q7PX28 A0A182L8F2 A0A182QU12 W5J189 A0A182PJ36 A0A182VW47 A0A182HK72 A0A182XP36 A0A0C9R126 Q175Y8 A0A151X8I1 A0A026WWM1 A0A154PEG1 K7J4Z0 A0A182YHT2 A0A0K8V0X3 A0A3B0KJA3 F4W7J6 B5DX93 B4GLN6 B4LXL6 W8BMP0 A0A195ESL3 A0A1B0A1P5 A0A0A1XSV9 A0A182GDR0 A0A1A9VL99 A0A195EL28 A0A1Q3FSJ8 A0A0M8ZZT7 B4NAX6 A0A2P8XTX8 A0A1A9X2N1 A0A0A1WU89 A0A2J7QJP6 Q16LC9 A0A1B0FN19 A0A1I8Q4W1 A0A1Q3FSH6 B4JTK2 A0A2A3EKX1 A0A067QX91 V9IKT4 A0A1B0BA90 A0A2J7QJQ3 W8BJ82 A0A0K8VUV5 A0A084W0F7 A0A232EYM7 A0A1Q3G536 B4K9T9 A0A1A9XW60 A0A195AT73 A0A1W4XEU6 A0A1W4X5A6 A0A2A4JF18 A0A182NQW1 A0A0L0CPB2 A0A1W4WCA6 T1GHV9 B3LXC3 A0A212FLL7 A0A1I8Q8H6 A0A1I8MDU3 A0A182PGU7 A0A1J1HJW9 A0A182VWJ0 A0A0M4F4Q2 A0A1Y9IMI8 A0A182L9J4 A0A182ULS2 A0A0L0C9P3 A0A182WSB4 A0A182HL66 E2A8C5 A0A182SGA4 T1PAX5 A0A1B6DZY1 Q7PXS9 A0A1I8MZG3 A0A034V935 A0A336KMW3 A0A182RSH4 A0A182JVH7 A0A182VI42 A0A182TWQ2 A0A1B0EY77 A0A1L8DET3 A0A1A9ZES2 A0A1B0G0B1 W5J3T7 A0A1A9VCT3 A0A1B6CDJ6 E0VCA6
PDB
5NVA
E-value=3.86944e-30,
Score=330
Ontologies
GO
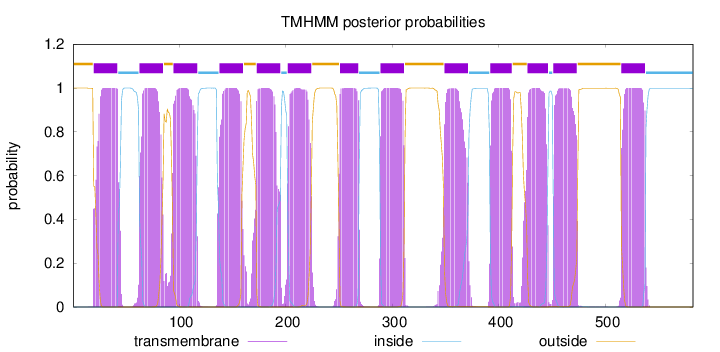
Topology
Length:
582
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
285.23935
Exp number, first 60 AAs:
22.39023
Total prob of N-in:
0.00032
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 62
TMhelix
63 - 85
outside
86 - 94
TMhelix
95 - 117
inside
118 - 137
TMhelix
138 - 160
outside
161 - 172
TMhelix
173 - 195
inside
196 - 201
TMhelix
202 - 224
outside
225 - 250
TMhelix
251 - 268
inside
269 - 288
TMhelix
289 - 311
outside
312 - 348
TMhelix
349 - 371
inside
372 - 391
TMhelix
392 - 412
outside
413 - 426
TMhelix
427 - 446
inside
447 - 450
TMhelix
451 - 473
outside
474 - 514
TMhelix
515 - 537
inside
538 - 582
Population Genetic Test Statistics
Pi
298.251112
Theta
233.315201
Tajima's D
0.082438
CLR
137.810915
CSRT
0.388330583470826
Interpretation
Uncertain
