Pre Gene Modal
BGIBMGA002671
Annotation
exosc7_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.625 Nuclear Reliability : 1.036
Sequence
CDS
ATGGCAGGACTGTTATTAAGTTCTACAGAAAAAGTTTTTATTGTTCACGGTGTACAAGAGGACTTTCGTTCCGATGGCCGATCAAACATTGACTATAGGCCCATGGAACTAGAAACAGATGTAGTGAGCCACGCAAGTGGCTCAGCGCGGCTTCGTCTAGCAAACACAGACATTTTAGTTGGCATTAAAACAGAAATAGATACACCAAAGCCAGACAAACCAGATATGGGAAAAATTGAGTTTTTTGTTGATTGCTCAGCAAATGCCACACCGGAATTCGAAGGGCGTGGTGGAGAAAATTTAGCTAGCAGCATTTCAAACACAATGCAAAGAGCATATCACTCATCACAAGCCTTTGATTTGAAGCAACTATGCATTGTAGAAGGCAAACAGTGCTGGAAACTTTATGTTGATATATTGATCTTGGAATGTGGGGGTAATTTGTTTGATGCAGTTTCATTAGCAACCAAAGCAGCTCTGTTTAACACAAGGATACCATTAGTAAAAGCCTTGTTGATGGATGGAGGAAATGTTGATTTACAACTCTCAGATGATCCTTATGATTTCAAATTTCTAGATGTTGGTTCTGCACCTTTGTTGGTAACGCTATGTAAAATAGGAGAAAAATGCGTGGTAGATCCCTCAGCTGAAGAAGAAAGCTGCAGTGTAATATCATTGATAGTTGGAATTACGGGAAACAGCAAATCTTACTGTGCAGATAAAGGAGCTTCTAACATAACTGATGTTGACATTAAATGTAGTACAATCAGTATGAACGGACCAGGCAGTTTAGCACCAAAGACATTAAAGAATGCAATTCACCAGGGCATCATTGCTGCAAAGGTACTAGATGATTCATTAGGACAAGCCTTACTGCGTGAACGACGAGAAAATGTCAACTCAATGAAAAATTCATATGGATTTCTTAAATAA
Protein
MAGLLLSSTEKVFIVHGVQEDFRSDGRSNIDYRPMELETDVVSHASGSARLRLANTDILVGIKTEIDTPKPDKPDMGKIEFFVDCSANATPEFEGRGGENLASSISNTMQRAYHSSQAFDLKQLCIVEGKQCWKLYVDILILECGGNLFDAVSLATKAALFNTRIPLVKALLMDGGNVDLQLSDDPYDFKFLDVGSAPLLVTLCKIGEKCVVDPSAEEESCSVISLIVGITGNSKSYCADKGASNITDVDIKCSTISMNGPGSLAPKTLKNAIHQGIIAAKVLDDSLGQALLRERRENVNSMKNSYGFLK
Summary
Uniprot
H9IZI6
Q1HPI0
A0A212FLM3
A0A194R765
A0A194Q2C3
S4PLS4
+ More
A0A2A4JGP3 A0A182J3L9 U5ES28 A0A182V8M8 A0A2J7R2R5 A0A1S4FBS3 A0A182IEQ6 Q7QDY7 Q178L5 A0A182GPW0 A0A182LW78 A0A0A1WYV6 A0A0K8VWM2 W8C6F5 A0A1Q3FXV9 A0A034W9Z5 A0A182NDE8 A0A182W3Y1 A0A182RJT9 A0A182QG01 A0A1B0DPE1 A0A1I8M9L1 A0A336LSB5 A0A1B6ID95 A0A1B6GX30 A0A1B6LN19 A0A1I8QEX9 A0A0L0CAS3 A0A182FBW6 A0A1B0CNM0 A0A1A9YUE2 A0A1L8DYA4 A0A1A9UI53 A0A182U895 W5JFK5 A0A182KZZ4 A0A1A9XG14 A0A1B0BGB2 A0A1B6CR00 A0A0M5JA58 A0A1A9WDX9 A0A0K8VG47 B4MQK0 D6WDU3 A0A084VUR0 B3MF61 B4KQC1 B4J9X0 A0A1W4X518 A0A1Y1LI02 B4LL43 A0A182T295 B4H534 A0A2M4BW12 A0A182PU26 Q28X67 A0A1J1IBM2 A0A182YIU7 B4P6S5 A0A1W4V0C5 A0A3B0J1P5 B3NPY6 C3XU17 A0A0L8G0U0 A0A1A9Z5L6 Q29QQ9 A0A2H8TH04 A0A210R233 A0A195E3P6 A0A0J9RDK3 A0A091DMC2 A0A154NX20 I3M6V5 A0A1Y1LHY3 F4WWV1 J9KAN7 F1SRC7 H0WHP2 A0A151I261 A0A158NKY3 A0A1U7UHG6 F7BW26 A0A2A3EI94 V9IC95 H0V4A8 E0VZC4 E9IZ26 A0A195FRU4 B2RDZ9 A0A099ZBX4 A0A2K6MET8
A0A2A4JGP3 A0A182J3L9 U5ES28 A0A182V8M8 A0A2J7R2R5 A0A1S4FBS3 A0A182IEQ6 Q7QDY7 Q178L5 A0A182GPW0 A0A182LW78 A0A0A1WYV6 A0A0K8VWM2 W8C6F5 A0A1Q3FXV9 A0A034W9Z5 A0A182NDE8 A0A182W3Y1 A0A182RJT9 A0A182QG01 A0A1B0DPE1 A0A1I8M9L1 A0A336LSB5 A0A1B6ID95 A0A1B6GX30 A0A1B6LN19 A0A1I8QEX9 A0A0L0CAS3 A0A182FBW6 A0A1B0CNM0 A0A1A9YUE2 A0A1L8DYA4 A0A1A9UI53 A0A182U895 W5JFK5 A0A182KZZ4 A0A1A9XG14 A0A1B0BGB2 A0A1B6CR00 A0A0M5JA58 A0A1A9WDX9 A0A0K8VG47 B4MQK0 D6WDU3 A0A084VUR0 B3MF61 B4KQC1 B4J9X0 A0A1W4X518 A0A1Y1LI02 B4LL43 A0A182T295 B4H534 A0A2M4BW12 A0A182PU26 Q28X67 A0A1J1IBM2 A0A182YIU7 B4P6S5 A0A1W4V0C5 A0A3B0J1P5 B3NPY6 C3XU17 A0A0L8G0U0 A0A1A9Z5L6 Q29QQ9 A0A2H8TH04 A0A210R233 A0A195E3P6 A0A0J9RDK3 A0A091DMC2 A0A154NX20 I3M6V5 A0A1Y1LHY3 F4WWV1 J9KAN7 F1SRC7 H0WHP2 A0A151I261 A0A158NKY3 A0A1U7UHG6 F7BW26 A0A2A3EI94 V9IC95 H0V4A8 E0VZC4 E9IZ26 A0A195FRU4 B2RDZ9 A0A099ZBX4 A0A2K6MET8
Pubmed
19121390
22118469
26354079
23622113
12364791
14747013
+ More
17210077 17510324 26483478 25830018 24495485 25348373 25315136 26108605 20920257 23761445 20966253 17994087 18362917 19820115 24438588 28004739 15632085 25244985 17550304 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 22936249 21719571 30723633 21347285 19892987 21993624 20566863 21282665
17210077 17510324 26483478 25830018 24495485 25348373 25315136 26108605 20920257 23761445 20966253 17994087 18362917 19820115 24438588 28004739 15632085 25244985 17550304 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 22936249 21719571 30723633 21347285 19892987 21993624 20566863 21282665
EMBL
BABH01014251
DQ443422
ABF51511.1
AGBW02007743
OWR54627.1
KQ460870
+ More
KPJ11711.1 KQ459589 KPI97555.1 GAIX01003880 JAA88680.1 NWSH01001649 PCG70562.1 GANO01002551 JAB57320.1 NEVH01007828 PNF35123.1 APCN01009333 AAAB01008849 EAA07136.2 CH477362 EAT42627.1 JXUM01016535 KQ560461 KXJ82305.1 AXCM01000614 GBXI01010689 JAD03603.1 GDHF01009344 JAI42970.1 GAMC01004289 JAC02267.1 GFDL01002712 JAV32333.1 GAKP01007428 JAC51524.1 AXCN02002125 AJVK01008029 UFQT01000086 UFQT01001813 SSX19469.1 SSX31887.1 GECU01022810 JAS84896.1 GECZ01002800 JAS66969.1 GEBQ01015083 JAT24894.1 JRES01000655 KNC29503.1 AJWK01020518 CCAG010004750 GFDF01002849 JAV11235.1 ADMH02001541 ETN62123.1 JXJN01013815 GEDC01021460 JAS15838.1 CP012524 ALC41306.1 GDHF01014501 JAI37813.1 CH963849 EDW74389.1 KQ971323 EEZ99918.1 ATLV01016988 KE525139 KFB41704.1 CH902619 EDV37689.1 CH933808 EDW09249.1 CH916367 EDW02557.1 GEZM01055002 JAV73263.1 CH940648 EDW61850.1 CH479210 EDW32870.1 GGFJ01008125 MBW57266.1 CM000071 EAL26449.1 CVRI01000047 CRK97679.1 CM000158 EDW92002.1 OUUW01000001 SPP74885.1 CH954179 EDV55833.1 GG666464 EEN68391.1 KQ424671 KOF70626.1 AE013599 BT024331 AAF58076.3 ABC86393.1 GFXV01001505 MBW13310.1 NEDP02000756 OWF55143.1 KQ979701 KYN19691.1 CM002911 KMY94143.1 KN122104 KFO33269.1 KQ434777 KZC04226.1 AGTP01095401 GEZM01055003 JAV73262.1 GL888413 EGI61339.1 ABLF02039764 ABLF02039778 AEMK02000085 DQIR01285683 HDC41161.1 AAQR03112320 AAQR03112321 AAQR03112322 AAQR03112323 AAQR03112324 KQ976546 KYM80939.1 ADTU01019028 KZ288253 PBC30992.1 JR038503 AEY58282.1 AAKN02012259 AAKN02012260 DS235851 EEB18730.1 GL767078 EFZ14187.1 KQ981281 KYN43325.1 AK315741 BAG38096.1 KL891119 KGL78473.1
KPJ11711.1 KQ459589 KPI97555.1 GAIX01003880 JAA88680.1 NWSH01001649 PCG70562.1 GANO01002551 JAB57320.1 NEVH01007828 PNF35123.1 APCN01009333 AAAB01008849 EAA07136.2 CH477362 EAT42627.1 JXUM01016535 KQ560461 KXJ82305.1 AXCM01000614 GBXI01010689 JAD03603.1 GDHF01009344 JAI42970.1 GAMC01004289 JAC02267.1 GFDL01002712 JAV32333.1 GAKP01007428 JAC51524.1 AXCN02002125 AJVK01008029 UFQT01000086 UFQT01001813 SSX19469.1 SSX31887.1 GECU01022810 JAS84896.1 GECZ01002800 JAS66969.1 GEBQ01015083 JAT24894.1 JRES01000655 KNC29503.1 AJWK01020518 CCAG010004750 GFDF01002849 JAV11235.1 ADMH02001541 ETN62123.1 JXJN01013815 GEDC01021460 JAS15838.1 CP012524 ALC41306.1 GDHF01014501 JAI37813.1 CH963849 EDW74389.1 KQ971323 EEZ99918.1 ATLV01016988 KE525139 KFB41704.1 CH902619 EDV37689.1 CH933808 EDW09249.1 CH916367 EDW02557.1 GEZM01055002 JAV73263.1 CH940648 EDW61850.1 CH479210 EDW32870.1 GGFJ01008125 MBW57266.1 CM000071 EAL26449.1 CVRI01000047 CRK97679.1 CM000158 EDW92002.1 OUUW01000001 SPP74885.1 CH954179 EDV55833.1 GG666464 EEN68391.1 KQ424671 KOF70626.1 AE013599 BT024331 AAF58076.3 ABC86393.1 GFXV01001505 MBW13310.1 NEDP02000756 OWF55143.1 KQ979701 KYN19691.1 CM002911 KMY94143.1 KN122104 KFO33269.1 KQ434777 KZC04226.1 AGTP01095401 GEZM01055003 JAV73262.1 GL888413 EGI61339.1 ABLF02039764 ABLF02039778 AEMK02000085 DQIR01285683 HDC41161.1 AAQR03112320 AAQR03112321 AAQR03112322 AAQR03112323 AAQR03112324 KQ976546 KYM80939.1 ADTU01019028 KZ288253 PBC30992.1 JR038503 AEY58282.1 AAKN02012259 AAKN02012260 DS235851 EEB18730.1 GL767078 EFZ14187.1 KQ981281 KYN43325.1 AK315741 BAG38096.1 KL891119 KGL78473.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000075880
+ More
UP000075903 UP000235965 UP000075840 UP000007062 UP000008820 UP000069940 UP000249989 UP000075883 UP000075884 UP000075920 UP000075900 UP000075886 UP000092462 UP000095301 UP000095300 UP000037069 UP000069272 UP000092461 UP000092444 UP000078200 UP000075902 UP000000673 UP000075882 UP000092443 UP000092460 UP000092553 UP000091820 UP000007798 UP000007266 UP000030765 UP000007801 UP000009192 UP000001070 UP000192223 UP000008792 UP000075901 UP000008744 UP000075885 UP000001819 UP000183832 UP000076408 UP000002282 UP000192221 UP000268350 UP000008711 UP000001554 UP000053454 UP000092445 UP000000803 UP000242188 UP000078492 UP000028990 UP000076502 UP000005215 UP000007755 UP000007819 UP000008227 UP000005225 UP000078540 UP000005205 UP000189704 UP000002281 UP000242457 UP000005447 UP000009046 UP000078541 UP000053641 UP000233180
UP000075903 UP000235965 UP000075840 UP000007062 UP000008820 UP000069940 UP000249989 UP000075883 UP000075884 UP000075920 UP000075900 UP000075886 UP000092462 UP000095301 UP000095300 UP000037069 UP000069272 UP000092461 UP000092444 UP000078200 UP000075902 UP000000673 UP000075882 UP000092443 UP000092460 UP000092553 UP000091820 UP000007798 UP000007266 UP000030765 UP000007801 UP000009192 UP000001070 UP000192223 UP000008792 UP000075901 UP000008744 UP000075885 UP000001819 UP000183832 UP000076408 UP000002282 UP000192221 UP000268350 UP000008711 UP000001554 UP000053454 UP000092445 UP000000803 UP000242188 UP000078492 UP000028990 UP000076502 UP000005215 UP000007755 UP000007819 UP000008227 UP000005225 UP000078540 UP000005205 UP000189704 UP000002281 UP000242457 UP000005447 UP000009046 UP000078541 UP000053641 UP000233180
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IZI6
Q1HPI0
A0A212FLM3
A0A194R765
A0A194Q2C3
S4PLS4
+ More
A0A2A4JGP3 A0A182J3L9 U5ES28 A0A182V8M8 A0A2J7R2R5 A0A1S4FBS3 A0A182IEQ6 Q7QDY7 Q178L5 A0A182GPW0 A0A182LW78 A0A0A1WYV6 A0A0K8VWM2 W8C6F5 A0A1Q3FXV9 A0A034W9Z5 A0A182NDE8 A0A182W3Y1 A0A182RJT9 A0A182QG01 A0A1B0DPE1 A0A1I8M9L1 A0A336LSB5 A0A1B6ID95 A0A1B6GX30 A0A1B6LN19 A0A1I8QEX9 A0A0L0CAS3 A0A182FBW6 A0A1B0CNM0 A0A1A9YUE2 A0A1L8DYA4 A0A1A9UI53 A0A182U895 W5JFK5 A0A182KZZ4 A0A1A9XG14 A0A1B0BGB2 A0A1B6CR00 A0A0M5JA58 A0A1A9WDX9 A0A0K8VG47 B4MQK0 D6WDU3 A0A084VUR0 B3MF61 B4KQC1 B4J9X0 A0A1W4X518 A0A1Y1LI02 B4LL43 A0A182T295 B4H534 A0A2M4BW12 A0A182PU26 Q28X67 A0A1J1IBM2 A0A182YIU7 B4P6S5 A0A1W4V0C5 A0A3B0J1P5 B3NPY6 C3XU17 A0A0L8G0U0 A0A1A9Z5L6 Q29QQ9 A0A2H8TH04 A0A210R233 A0A195E3P6 A0A0J9RDK3 A0A091DMC2 A0A154NX20 I3M6V5 A0A1Y1LHY3 F4WWV1 J9KAN7 F1SRC7 H0WHP2 A0A151I261 A0A158NKY3 A0A1U7UHG6 F7BW26 A0A2A3EI94 V9IC95 H0V4A8 E0VZC4 E9IZ26 A0A195FRU4 B2RDZ9 A0A099ZBX4 A0A2K6MET8
A0A2A4JGP3 A0A182J3L9 U5ES28 A0A182V8M8 A0A2J7R2R5 A0A1S4FBS3 A0A182IEQ6 Q7QDY7 Q178L5 A0A182GPW0 A0A182LW78 A0A0A1WYV6 A0A0K8VWM2 W8C6F5 A0A1Q3FXV9 A0A034W9Z5 A0A182NDE8 A0A182W3Y1 A0A182RJT9 A0A182QG01 A0A1B0DPE1 A0A1I8M9L1 A0A336LSB5 A0A1B6ID95 A0A1B6GX30 A0A1B6LN19 A0A1I8QEX9 A0A0L0CAS3 A0A182FBW6 A0A1B0CNM0 A0A1A9YUE2 A0A1L8DYA4 A0A1A9UI53 A0A182U895 W5JFK5 A0A182KZZ4 A0A1A9XG14 A0A1B0BGB2 A0A1B6CR00 A0A0M5JA58 A0A1A9WDX9 A0A0K8VG47 B4MQK0 D6WDU3 A0A084VUR0 B3MF61 B4KQC1 B4J9X0 A0A1W4X518 A0A1Y1LI02 B4LL43 A0A182T295 B4H534 A0A2M4BW12 A0A182PU26 Q28X67 A0A1J1IBM2 A0A182YIU7 B4P6S5 A0A1W4V0C5 A0A3B0J1P5 B3NPY6 C3XU17 A0A0L8G0U0 A0A1A9Z5L6 Q29QQ9 A0A2H8TH04 A0A210R233 A0A195E3P6 A0A0J9RDK3 A0A091DMC2 A0A154NX20 I3M6V5 A0A1Y1LHY3 F4WWV1 J9KAN7 F1SRC7 H0WHP2 A0A151I261 A0A158NKY3 A0A1U7UHG6 F7BW26 A0A2A3EI94 V9IC95 H0V4A8 E0VZC4 E9IZ26 A0A195FRU4 B2RDZ9 A0A099ZBX4 A0A2K6MET8
PDB
2NN6
E-value=5.21033e-81,
Score=765
Ontologies
GO
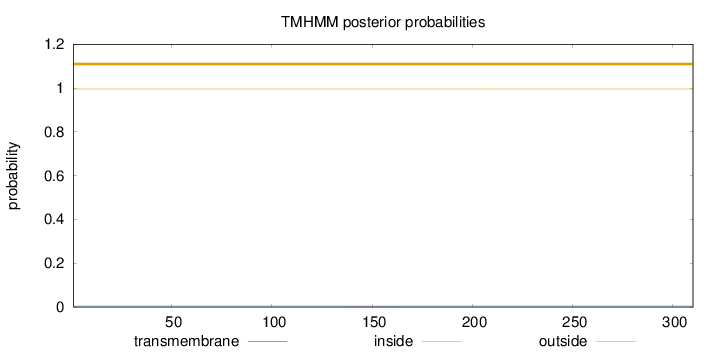
Topology
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00929999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00431
outside
1 - 310
Population Genetic Test Statistics
Pi
205.013388
Theta
190.338006
Tajima's D
0.850105
CLR
0.070117
CSRT
0.617369131543423
Interpretation
Uncertain
