Gene
KWMTBOMO02581
Pre Gene Modal
BGIBMGA002673
Annotation
PREDICTED:_arf-GAP_with_SH3_domain?_ANK_repeat_and_PH_domain-containing_protein_2_isoform_X2_[Bombyx_mori]
Full name
ArfGAP with SH3 domain, ANK repeat and PH domain-containing protein
Location in the cell
Nuclear Reliability : 3.853
Sequence
CDS
ATGCCGGGTCTTATTGGTATTGGTGAATTCATTGAGGAAACTCGAGAGGATTACAGTTCACCGACTACATCTACGTTCGTGTCACGAATTCCACAATGTCGTCAAACAATCAACGCTCTAGAAGAGACACTGGATTACGACCGCGATGGCCTAACGAAACTGAAGAAGGCGATAAAAGCCATACACAATTCCGGAAATGCCCACGTGGACAACGAGATGTATCTTTCACGAACACTGGAGCGGCTCGCTGGGAACGCTCTTAGCAAAGATTCCGAGCCAGATATTGGAGCAGCCTTCTTCAAGTTCGCTGTCGTCACAAAGGAACTATCGGCACTTATGAAGACTCTGATGCAAAACATTAACAACATAGTGATGTTTCCGGTCGATAGCCTTCTAAAAGGAGATCTGCGTGGTGTCAAAGGTGATCTAAAAAGGCCTTTCGACAAAGCTTCCAAAGATTACGAGTCTAAATATTTGAAAATCGAGAAAGAGAAAAAATCGCAAGCAAAAGAGGCTGGCCTCATACGTAGCGAAGTTACGTCCGCTGAAATAGCCGATGAAATGGAAAAAGAAAGGAGACTTTTTCAACTACAGATGTGCGAGTACCTGATCAAGTTCAATGAAATAAAAACCAAAAAAGGAATCGAGTTGCTTCAGCATTTAGTTGAGTACTATCACGCGCAGACAAATTATTTTCAAGACGGCTTGAAGACAATCGAACATTTCGGTGTTTATGTAGCTGATCTGAGTGTCCAGCTACAGAAAATTAGACAGCAACAAGACGACGAGAGAAGGGGGCTCCTAGAATTGAGGAACATGTTGAGGGCAGCAGCGCCACAGGATCGAGAGGTAGCCTCTTCGGTGGGAGGCTATTCTCTCCACCAACTGCAAGGCGATAAACAACACGGGGTGACCAGAAGTGGTTACCTTTTAAAAAAGTCCGAAGGTAAAGTAAGACGTGTTTGGCAAAAACGTCGCTGTCGGGTCGCCGCTGAAGGTTTTCTGGATATATTCCATGCCGATGAAAACAAGACTCCAGCTAGAGTTAATCTATTAACGTGCCAAATCAAAGTAGCCGCAGACGATAAGAGGGCGTTCGATCTAGTCTCATACAACCGTACCTATCACTTCCAAGCCGAAGACGAGGCCGAGCAAAGGGCTTGGACATCAGTTCTGGTGAACTGCAAGGAGGGGGCCCTGATGAGAGCGTTCCACCAGCAAGTAGGCGAGAGTAGTGACTCCGGTCATTCACTGCTAGATCTACAGAGATCTATTATACGCGCCGTTAGATCAATGCCCGGAAATCAAGTTTGCGCTGATTGCGGTTCGACAAATGATCCGACATGGCTATCGACAAATTTCGGTATAATAGTTTGTATCGAATGTTCTGGAAGTCACCGAGAACTCGGCGTTCATATATCGAGGATTCAATCTTTGACTTTAGATAGACTGAGCACTTCGCAGCTATTACTGGCCAGGAACATGGGAAATCAGATGTTCAATGAAGTCATGGAGAACACGCTCGATGATAGAGACAAACTCACGCCCGAGAGTACGATGGAAGAGCGTCTCAGGTTCATACGCGAGAAGTACGTGTACAGAGCGTGGGCGGCGCGCACGTGTCGCGACGCGGCCGAGCGTCTCTCCGAAGTCGAGCACGCAGTCAACAACGGACACTTGCAGAACCTGTTGCAGGCGTACGCTGAAGGCGCCGATCTGGCTGCGACGCTTCCTTGTTCGGACTGCGGCGAGACAGCCCTCCACTTGGCCATATCGCGGGAACTGGGCGACGGTTCCTGCTTGCACATAGTCGATTTTCTAGTCCAGAACGGTGGTTCTCAACTGTTAGACAAGGCGACCTCGACCGGCATGACGGCTCTGCATCTGTGCGCGGCGACCGATCGAACGGAACCGATGAAGTTGCTTCTCAAAGCCGGCGCCGATTCGTCGGTCAGGGACGGCAGCGGTCGGACGCCCTTGCAGATAGCGAGGCAGCTGGGCCATCATGGATGTCAGGAATTGTTGGAGAGTGTGGATAAGAGAGATAAAAACATATTCGAGAACATAAATATAGACTGGAACTTATCGCACGACGACGGCTCAACTGACTTCTCGGATGACGACACTGTTATCGACGAAAGACAAAACGGTAGCGTTACTCCGGAGAAGAAGTGTCCCCGTTCCCGGCCACCGTCGTACGCGGGTGGGTCGGTGGGTCCGGGCGCGGCGCCCGCGGGCAGCGCCGGCGGCGACTCCCCCGTGCTGCGGTCCCGCTCGTCGACCTGCGATTCGTTACACCACGCCCCCCCGACGCCAACTACGCTGCCAAGGAAACCTAACGTCTATAACATAGGCAGCTTAAAGAAGCGAGCGGCGCCCCTGCCCCCCGCCGTGGCGCTGTCCCACCCCCCGCCGCACCCCCGCTCTACTCCCTCGCCCTCCGCCGACACCACACGTTCACTTCACGTTATCAGCACAGCTTCGGGTGTGAAGCATCGTCCCCAGCAGCCTCCCCCCACCCCGCCGCCGACCGTCTCCTCGCTGCACAACGGCGCGTTTCACCGCGACGAACCGACGCCACCTCCCAGGAAGGAATGTTCTAGTCAAGACTCGTCATTAGAGTTGTGCGATATAACGGATTCGTGCTTGGACGAGAGTCGGTTGCATTCGGCCATGTCGTGCGACAGCTCGCGGTCCAGGGACCGATCCAGGCGCAGCGACAGTCGGTCGCTCGACGTCTCCGACACGTCCAGCGTGCACTCCAGATCGCCGTCTGCCTCTATCACTATGATGGGCGGCGTCAGACGGTGTCGGGCTCTGTACGATTGTTCGGCGGACAACGAAGACGAGCTGTCGTTCCGCGAGGGCGAAGTGATCGTGGTCATCAACGAACGCACCGAGGACGACAACTGGATGGAAGGTCAAGTCGAGGGCACGAGCAGGCGAGGGATGTTCCCCGTCTCTTTCGTTCACATGCTACCCGACTAG
Protein
MPGLIGIGEFIEETREDYSSPTTSTFVSRIPQCRQTINALEETLDYDRDGLTKLKKAIKAIHNSGNAHVDNEMYLSRTLERLAGNALSKDSEPDIGAAFFKFAVVTKELSALMKTLMQNINNIVMFPVDSLLKGDLRGVKGDLKRPFDKASKDYESKYLKIEKEKKSQAKEAGLIRSEVTSAEIADEMEKERRLFQLQMCEYLIKFNEIKTKKGIELLQHLVEYYHAQTNYFQDGLKTIEHFGVYVADLSVQLQKIRQQQDDERRGLLELRNMLRAAAPQDREVASSVGGYSLHQLQGDKQHGVTRSGYLLKKSEGKVRRVWQKRRCRVAAEGFLDIFHADENKTPARVNLLTCQIKVAADDKRAFDLVSYNRTYHFQAEDEAEQRAWTSVLVNCKEGALMRAFHQQVGESSDSGHSLLDLQRSIIRAVRSMPGNQVCADCGSTNDPTWLSTNFGIIVCIECSGSHRELGVHISRIQSLTLDRLSTSQLLLARNMGNQMFNEVMENTLDDRDKLTPESTMEERLRFIREKYVYRAWAARTCRDAAERLSEVEHAVNNGHLQNLLQAYAEGADLAATLPCSDCGETALHLAISRELGDGSCLHIVDFLVQNGGSQLLDKATSTGMTALHLCAATDRTEPMKLLLKAGADSSVRDGSGRTPLQIARQLGHHGCQELLESVDKRDKNIFENINIDWNLSHDDGSTDFSDDDTVIDERQNGSVTPEKKCPRSRPPSYAGGSVGPGAAPAGSAGGDSPVLRSRSSTCDSLHHAPPTPTTLPRKPNVYNIGSLKKRAAPLPPAVALSHPPPHPRSTPSPSADTTRSLHVISTASGVKHRPQQPPPTPPPTVSSLHNGAFHRDEPTPPPRKECSSQDSSLELCDITDSCLDESRLHSAMSCDSSRSRDRSRRSDSRSLDVSDTSSVHSRSPSASITMMGGVRRCRALYDCSADNEDELSFREGEVIVVINERTEDDNWMEGQVEGTSRRGMFPVSFVHMLPD
Summary
Description
Probable GTPase-activating protein (GAP) for Arf family proteins (Probable). Involved in Golgi apparatus organization by targeting Arf1 to the Golgi, which may be important for membrane trafficking during epithelial morphogenesis (PubMed:27535433). Regulates the positioning of interommatidial precursor cells during compound eye morphogenesis together with Arf6 and Cindr (PubMed:21976699). Required for cleavage furrow ingression in early embryonic cells (PubMed:27535433).
Subunit
Interacts with Cindr.
Keywords
Alternative splicing
ANK repeat
Cell membrane
Cell projection
Complete proteome
Cytoplasm
Developmental protein
GTPase activation
Membrane
Metal-binding
Nucleus
Reference proteome
Repeat
SH3 domain
Zinc
Zinc-finger
Feature
chain ArfGAP with SH3 domain, ANK repeat and PH domain-containing protein
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A212FLQ7
A0A194PXS9
A0A194R397
A0A2A4J5X3
A0A2A4J4M8
A0A2H1WAI5
+ More
A0A0L7L7N6 A0A195C3T7 A0A195ELZ5 A0A195B6H5 A0A195ESR4 A0A026VW10 A0A3L8D5R0 A0A088AAB7 A0A154NZS6 A0A1B6DAJ4 A0A232EF41 K7IPS3 A0A1B6D7S1 A0A023EWH3 A0A067R5P9 A0A1B6CR36 A0A1Y1KEF8 T1HS89 A0A1B6C689 E0VXN3 A0A1W4X2F8 A0A1W4WST7 A0A0A9ZB56 A0A1B6ISM5 A0A1B6J1N0 A0A0A9Z888 A0A1Y1KAX0 A0A1B6H7G1 A0A2A3E3S4 A0A2M4AC38 A0A2M4ABY7 A0A182FFZ0 A0A0M9AB16 W5JC19 A0A1L8DMM7 A0A1L8DMS1 A0A1L8DMI6 A0A1L8DMH0 A0A182J4U7 A0A182MRF2 A0A182KS20 A7UR68 A0A182SNB7 Q7QJY3 A0A182WJ09 A0A182NHA1 A0A182QM39 A0A182RB02 A0A2J7RBA1 A0A182XZA0 W8B3C3 A0A182I3Y1 W8AZ75 W8ART4 A0A0K8W7F4 A0A0K8U2F6 A0A0A1XHL2 A0A1J1J935 A0A0K8W541 A0A0K8VH20 A0A0K8TZH1 A0A0K8VDK0 A0A034VN40 A0A3B0JPG8 A0A0Q9X7T1 A0A0M3QUF3 A0A1W4VZA8 A0A1W4WA47 A0A0J9R801 A0A0J9TW00 A0A0Q9X8M7 Q28ZW8 A0A0R3NSM5 A0A0P8Y6M9 B3MGE5 A1Z7A6 A0A0Q9XJC0 B4KNX1 B3N985 B4P292 D6W8A3 A0A0J9R934 A1Z7A6-2 A0A1W4WB25 A0A0J9R7S9 B4GGN8 A0A0B4LEF8 A0A0P8ZRJ3 A0A1W4WBI2 A0A0Q9W4U2 A0A0P8XR67 B4LJM2 A0A0R1DRI8 B4NMJ1
A0A0L7L7N6 A0A195C3T7 A0A195ELZ5 A0A195B6H5 A0A195ESR4 A0A026VW10 A0A3L8D5R0 A0A088AAB7 A0A154NZS6 A0A1B6DAJ4 A0A232EF41 K7IPS3 A0A1B6D7S1 A0A023EWH3 A0A067R5P9 A0A1B6CR36 A0A1Y1KEF8 T1HS89 A0A1B6C689 E0VXN3 A0A1W4X2F8 A0A1W4WST7 A0A0A9ZB56 A0A1B6ISM5 A0A1B6J1N0 A0A0A9Z888 A0A1Y1KAX0 A0A1B6H7G1 A0A2A3E3S4 A0A2M4AC38 A0A2M4ABY7 A0A182FFZ0 A0A0M9AB16 W5JC19 A0A1L8DMM7 A0A1L8DMS1 A0A1L8DMI6 A0A1L8DMH0 A0A182J4U7 A0A182MRF2 A0A182KS20 A7UR68 A0A182SNB7 Q7QJY3 A0A182WJ09 A0A182NHA1 A0A182QM39 A0A182RB02 A0A2J7RBA1 A0A182XZA0 W8B3C3 A0A182I3Y1 W8AZ75 W8ART4 A0A0K8W7F4 A0A0K8U2F6 A0A0A1XHL2 A0A1J1J935 A0A0K8W541 A0A0K8VH20 A0A0K8TZH1 A0A0K8VDK0 A0A034VN40 A0A3B0JPG8 A0A0Q9X7T1 A0A0M3QUF3 A0A1W4VZA8 A0A1W4WA47 A0A0J9R801 A0A0J9TW00 A0A0Q9X8M7 Q28ZW8 A0A0R3NSM5 A0A0P8Y6M9 B3MGE5 A1Z7A6 A0A0Q9XJC0 B4KNX1 B3N985 B4P292 D6W8A3 A0A0J9R934 A1Z7A6-2 A0A1W4WB25 A0A0J9R7S9 B4GGN8 A0A0B4LEF8 A0A0P8ZRJ3 A0A1W4WBI2 A0A0Q9W4U2 A0A0P8XR67 B4LJM2 A0A0R1DRI8 B4NMJ1
Pubmed
22118469
26354079
26227816
24508170
30249741
28648823
+ More
20075255 25474469 24845553 28004739 20566863 25401762 20920257 23761445 20966253 12364791 25244985 24495485 25830018 25348373 17994087 22936249 15632085 10731132 12537572 12537569 21976699 27535433 17550304 18362917 19820115 12537568 12537573 12537574 16110336 17569856 17569867
20075255 25474469 24845553 28004739 20566863 25401762 20920257 23761445 20966253 12364791 25244985 24495485 25830018 25348373 17994087 22936249 15632085 10731132 12537572 12537569 21976699 27535433 17550304 18362917 19820115 12537568 12537573 12537574 16110336 17569856 17569867
EMBL
AGBW02007743
OWR54630.1
KQ459589
KPI97559.1
KQ460870
KPJ11715.1
+ More
NWSH01003164 PCG66840.1 PCG66839.1 ODYU01007382 SOQ50091.1 JTDY01002469 KOB71354.1 KQ978292 KYM95524.1 KQ978739 KYN28899.1 KQ976582 KYM79860.1 KQ981979 KYN31273.1 KK107796 EZA47681.1 QOIP01000013 RLU15353.1 KQ434786 KZC05127.1 GEDC01014585 JAS22713.1 NNAY01005161 OXU16932.1 GEDC01015576 JAS21722.1 GBBI01005057 JAC13655.1 KK852945 KDR13464.1 GEDC01021434 JAS15864.1 GEZM01087598 JAV58590.1 ACPB03009798 GEDC01028311 JAS08987.1 DS235833 EEB18139.1 GBHO01036884 GBHO01003016 JAG06720.1 JAG40588.1 GECU01017775 JAS89931.1 GECU01014634 JAS93072.1 GBHO01005564 GBHO01005563 JAG38040.1 JAG38041.1 GEZM01087599 JAV58589.1 GECU01037041 JAS70665.1 KZ288392 PBC26338.1 GGFK01005026 MBW38347.1 GGFK01004976 MBW38297.1 KQ435695 KOX80867.1 ADMH02001644 ETN61631.1 GFDF01006457 JAV07627.1 GFDF01006420 JAV07664.1 GFDF01006422 JAV07662.1 GFDF01006421 JAV07663.1 AXCM01012044 AAAB01008807 EDO64493.1 EAA04701.3 AXCN02000894 NEVH01005921 PNF38102.1 GAMC01015017 JAB91538.1 APCN01000228 GAMC01015018 JAB91537.1 GAMC01015015 JAB91540.1 GDHF01005248 GDHF01000856 JAI47066.1 JAI51458.1 GDHF01031794 GDHF01022911 JAI20520.1 JAI29403.1 GBXI01005947 GBXI01003887 JAD08345.1 JAD10405.1 CVRI01000075 CRL08570.1 GDHF01006135 GDHF01004200 GDHF01002156 JAI46179.1 JAI48114.1 JAI50158.1 GDHF01029060 GDHF01014110 JAI23254.1 JAI38204.1 GDHF01032440 GDHF01029582 JAI19874.1 JAI22732.1 GDHF01024342 GDHF01021086 GDHF01015295 GDHF01015066 JAI27972.1 JAI31228.1 JAI37019.1 JAI37248.1 GAKP01014247 GAKP01014246 JAC44706.1 OUUW01000001 SPP75236.1 CH933808 KRG04422.1 CP012524 ALC40563.1 CM002911 KMY92191.1 KMY92190.1 KRG04421.1 CM000071 EAL25494.2 KRT02271.1 CH902619 KPU77056.1 EDV37848.1 AE013599 BT015252 BT031167 AY061577 AAL29125.1 AAT94481.1 ABX00789.1 KRG04423.1 EDW09016.1 CH954177 EDV59572.1 CM000157 EDW89293.1 KQ971307 EFA10890.2 KMY92194.1 KMY92192.1 CH479183 EDW35658.1 AHN55981.1 KPU77055.1 CH940648 KRF80090.1 KPU77057.1 EDW61590.2 KRJ98354.1 CH964282 EDW85580.2
NWSH01003164 PCG66840.1 PCG66839.1 ODYU01007382 SOQ50091.1 JTDY01002469 KOB71354.1 KQ978292 KYM95524.1 KQ978739 KYN28899.1 KQ976582 KYM79860.1 KQ981979 KYN31273.1 KK107796 EZA47681.1 QOIP01000013 RLU15353.1 KQ434786 KZC05127.1 GEDC01014585 JAS22713.1 NNAY01005161 OXU16932.1 GEDC01015576 JAS21722.1 GBBI01005057 JAC13655.1 KK852945 KDR13464.1 GEDC01021434 JAS15864.1 GEZM01087598 JAV58590.1 ACPB03009798 GEDC01028311 JAS08987.1 DS235833 EEB18139.1 GBHO01036884 GBHO01003016 JAG06720.1 JAG40588.1 GECU01017775 JAS89931.1 GECU01014634 JAS93072.1 GBHO01005564 GBHO01005563 JAG38040.1 JAG38041.1 GEZM01087599 JAV58589.1 GECU01037041 JAS70665.1 KZ288392 PBC26338.1 GGFK01005026 MBW38347.1 GGFK01004976 MBW38297.1 KQ435695 KOX80867.1 ADMH02001644 ETN61631.1 GFDF01006457 JAV07627.1 GFDF01006420 JAV07664.1 GFDF01006422 JAV07662.1 GFDF01006421 JAV07663.1 AXCM01012044 AAAB01008807 EDO64493.1 EAA04701.3 AXCN02000894 NEVH01005921 PNF38102.1 GAMC01015017 JAB91538.1 APCN01000228 GAMC01015018 JAB91537.1 GAMC01015015 JAB91540.1 GDHF01005248 GDHF01000856 JAI47066.1 JAI51458.1 GDHF01031794 GDHF01022911 JAI20520.1 JAI29403.1 GBXI01005947 GBXI01003887 JAD08345.1 JAD10405.1 CVRI01000075 CRL08570.1 GDHF01006135 GDHF01004200 GDHF01002156 JAI46179.1 JAI48114.1 JAI50158.1 GDHF01029060 GDHF01014110 JAI23254.1 JAI38204.1 GDHF01032440 GDHF01029582 JAI19874.1 JAI22732.1 GDHF01024342 GDHF01021086 GDHF01015295 GDHF01015066 JAI27972.1 JAI31228.1 JAI37019.1 JAI37248.1 GAKP01014247 GAKP01014246 JAC44706.1 OUUW01000001 SPP75236.1 CH933808 KRG04422.1 CP012524 ALC40563.1 CM002911 KMY92191.1 KMY92190.1 KRG04421.1 CM000071 EAL25494.2 KRT02271.1 CH902619 KPU77056.1 EDV37848.1 AE013599 BT015252 BT031167 AY061577 AAL29125.1 AAT94481.1 ABX00789.1 KRG04423.1 EDW09016.1 CH954177 EDV59572.1 CM000157 EDW89293.1 KQ971307 EFA10890.2 KMY92194.1 KMY92192.1 CH479183 EDW35658.1 AHN55981.1 KPU77055.1 CH940648 KRF80090.1 KPU77057.1 EDW61590.2 KRJ98354.1 CH964282 EDW85580.2
Proteomes
UP000007151
UP000053268
UP000053240
UP000218220
UP000037510
UP000078542
+ More
UP000078492 UP000078540 UP000078541 UP000053097 UP000279307 UP000005203 UP000076502 UP000215335 UP000002358 UP000027135 UP000015103 UP000009046 UP000192223 UP000242457 UP000069272 UP000053105 UP000000673 UP000075880 UP000075883 UP000075882 UP000007062 UP000075901 UP000075920 UP000075884 UP000075886 UP000075900 UP000235965 UP000076408 UP000075840 UP000183832 UP000268350 UP000009192 UP000092553 UP000192221 UP000001819 UP000007801 UP000000803 UP000008711 UP000002282 UP000007266 UP000008744 UP000008792 UP000007798
UP000078492 UP000078540 UP000078541 UP000053097 UP000279307 UP000005203 UP000076502 UP000215335 UP000002358 UP000027135 UP000015103 UP000009046 UP000192223 UP000242457 UP000069272 UP000053105 UP000000673 UP000075880 UP000075883 UP000075882 UP000007062 UP000075901 UP000075920 UP000075884 UP000075886 UP000075900 UP000235965 UP000076408 UP000075840 UP000183832 UP000268350 UP000009192 UP000092553 UP000192221 UP000001819 UP000007801 UP000000803 UP000008711 UP000002282 UP000007266 UP000008744 UP000008792 UP000007798
PRIDE
Pfam
Interpro
IPR036028
SH3-like_dom_sf
+ More
IPR001849 PH_domain
IPR037278 ARFGAP/RecO
IPR002110 Ankyrin_rpt
IPR011993 PH-like_dom_sf
IPR000679 Znf_GATA
IPR020683 Ankyrin_rpt-contain_dom
IPR001164 ArfGAP_dom
IPR027267 AH/BAR_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR001452 SH3_domain
IPR037844 PH_ASAP
IPR038508 ArfGAP_dom_sf
IPR035836 ASAP1-like_SH3
IPR018225 Transaldolase_AS
IPR001012 UBX_dom
IPR012989 SEP_domain
IPR011249 Metalloenz_LuxS/M16
IPR029044 Nucleotide-diphossugar_trans
IPR013578 Peptidase_M16C_assoc
IPR009060 UBA-like_sf
IPR005027 Glyco_trans_43
IPR036241 NSFL1C_SEP_dom_sf
IPR029071 Ubiquitin-like_domsf
IPR007863 Peptidase_M16_C
IPR001849 PH_domain
IPR037278 ARFGAP/RecO
IPR002110 Ankyrin_rpt
IPR011993 PH-like_dom_sf
IPR000679 Znf_GATA
IPR020683 Ankyrin_rpt-contain_dom
IPR001164 ArfGAP_dom
IPR027267 AH/BAR_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR001452 SH3_domain
IPR037844 PH_ASAP
IPR038508 ArfGAP_dom_sf
IPR035836 ASAP1-like_SH3
IPR018225 Transaldolase_AS
IPR001012 UBX_dom
IPR012989 SEP_domain
IPR011249 Metalloenz_LuxS/M16
IPR029044 Nucleotide-diphossugar_trans
IPR013578 Peptidase_M16C_assoc
IPR009060 UBA-like_sf
IPR005027 Glyco_trans_43
IPR036241 NSFL1C_SEP_dom_sf
IPR029071 Ubiquitin-like_domsf
IPR007863 Peptidase_M16_C
SUPFAM
ProteinModelPortal
A0A212FLQ7
A0A194PXS9
A0A194R397
A0A2A4J5X3
A0A2A4J4M8
A0A2H1WAI5
+ More
A0A0L7L7N6 A0A195C3T7 A0A195ELZ5 A0A195B6H5 A0A195ESR4 A0A026VW10 A0A3L8D5R0 A0A088AAB7 A0A154NZS6 A0A1B6DAJ4 A0A232EF41 K7IPS3 A0A1B6D7S1 A0A023EWH3 A0A067R5P9 A0A1B6CR36 A0A1Y1KEF8 T1HS89 A0A1B6C689 E0VXN3 A0A1W4X2F8 A0A1W4WST7 A0A0A9ZB56 A0A1B6ISM5 A0A1B6J1N0 A0A0A9Z888 A0A1Y1KAX0 A0A1B6H7G1 A0A2A3E3S4 A0A2M4AC38 A0A2M4ABY7 A0A182FFZ0 A0A0M9AB16 W5JC19 A0A1L8DMM7 A0A1L8DMS1 A0A1L8DMI6 A0A1L8DMH0 A0A182J4U7 A0A182MRF2 A0A182KS20 A7UR68 A0A182SNB7 Q7QJY3 A0A182WJ09 A0A182NHA1 A0A182QM39 A0A182RB02 A0A2J7RBA1 A0A182XZA0 W8B3C3 A0A182I3Y1 W8AZ75 W8ART4 A0A0K8W7F4 A0A0K8U2F6 A0A0A1XHL2 A0A1J1J935 A0A0K8W541 A0A0K8VH20 A0A0K8TZH1 A0A0K8VDK0 A0A034VN40 A0A3B0JPG8 A0A0Q9X7T1 A0A0M3QUF3 A0A1W4VZA8 A0A1W4WA47 A0A0J9R801 A0A0J9TW00 A0A0Q9X8M7 Q28ZW8 A0A0R3NSM5 A0A0P8Y6M9 B3MGE5 A1Z7A6 A0A0Q9XJC0 B4KNX1 B3N985 B4P292 D6W8A3 A0A0J9R934 A1Z7A6-2 A0A1W4WB25 A0A0J9R7S9 B4GGN8 A0A0B4LEF8 A0A0P8ZRJ3 A0A1W4WBI2 A0A0Q9W4U2 A0A0P8XR67 B4LJM2 A0A0R1DRI8 B4NMJ1
A0A0L7L7N6 A0A195C3T7 A0A195ELZ5 A0A195B6H5 A0A195ESR4 A0A026VW10 A0A3L8D5R0 A0A088AAB7 A0A154NZS6 A0A1B6DAJ4 A0A232EF41 K7IPS3 A0A1B6D7S1 A0A023EWH3 A0A067R5P9 A0A1B6CR36 A0A1Y1KEF8 T1HS89 A0A1B6C689 E0VXN3 A0A1W4X2F8 A0A1W4WST7 A0A0A9ZB56 A0A1B6ISM5 A0A1B6J1N0 A0A0A9Z888 A0A1Y1KAX0 A0A1B6H7G1 A0A2A3E3S4 A0A2M4AC38 A0A2M4ABY7 A0A182FFZ0 A0A0M9AB16 W5JC19 A0A1L8DMM7 A0A1L8DMS1 A0A1L8DMI6 A0A1L8DMH0 A0A182J4U7 A0A182MRF2 A0A182KS20 A7UR68 A0A182SNB7 Q7QJY3 A0A182WJ09 A0A182NHA1 A0A182QM39 A0A182RB02 A0A2J7RBA1 A0A182XZA0 W8B3C3 A0A182I3Y1 W8AZ75 W8ART4 A0A0K8W7F4 A0A0K8U2F6 A0A0A1XHL2 A0A1J1J935 A0A0K8W541 A0A0K8VH20 A0A0K8TZH1 A0A0K8VDK0 A0A034VN40 A0A3B0JPG8 A0A0Q9X7T1 A0A0M3QUF3 A0A1W4VZA8 A0A1W4WA47 A0A0J9R801 A0A0J9TW00 A0A0Q9X8M7 Q28ZW8 A0A0R3NSM5 A0A0P8Y6M9 B3MGE5 A1Z7A6 A0A0Q9XJC0 B4KNX1 B3N985 B4P292 D6W8A3 A0A0J9R934 A1Z7A6-2 A0A1W4WB25 A0A0J9R7S9 B4GGN8 A0A0B4LEF8 A0A0P8ZRJ3 A0A1W4WBI2 A0A0Q9W4U2 A0A0P8XR67 B4LJM2 A0A0R1DRI8 B4NMJ1
PDB
1DCQ
E-value=3.60791e-62,
Score=608
Ontologies
GO
GO:0005096
GO:0006355
GO:0043565
GO:0008270
GO:0046872
GO:0005975
GO:0016021
GO:0016020
GO:0015018
GO:0006508
GO:1990386
GO:0005886
GO:0001745
GO:0005912
GO:0045177
GO:0005634
GO:0005737
GO:1903358
GO:1903829
GO:0043087
GO:0005902
GO:1901981
GO:0003700
GO:0005515
GO:0006631
GO:0006635
GO:0006281
GO:0048384
GO:0043547
Topology
Subcellular location
Cytoplasm
Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Nucleus Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Cell membrane Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Cell projection Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Microvillus Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Nucleus Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Cell membrane Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Cell projection Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
Microvillus Detected in large puncta at the plasma membrane (PubMed:21976699, PubMed:27535433). Excluded from the nucleus at interphase (PubMed:27535433). Enriched in the nucleus at prometaphase (PubMed:27535433). With evidence from 5 publications.
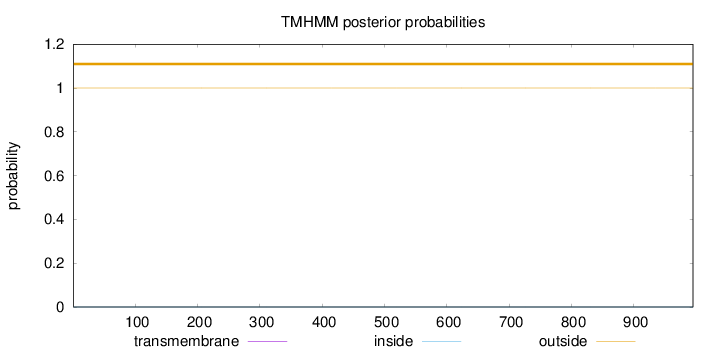
Length:
995
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9e-05
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 995
Population Genetic Test Statistics
Pi
264.9241
Theta
191.64388
Tajima's D
1.152266
CLR
0.183084
CSRT
0.693115344232788
Interpretation
Uncertain
