Gene
KWMTBOMO02580 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002674
Annotation
PREDICTED:_probable_hydroxyacid-oxoacid_transhydrogenase?_mitochondrial_[Bombyx_mori]
Full name
Probable hydroxyacid-oxoacid transhydrogenase, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.046 Mitochondrial Reliability : 1.419
Sequence
CDS
ATGACATCAAGAAAACGAATTTTCGATTTATTGCGAACGATAAATATTGCATCTTGCCAATGTCCTGCGCACGGTTACAGAGGAAATATTCAAGTTTCAAATGCAACACCATTAAAAGATTATGCATTTGAGATCAAATGCTCAACAGTTCGATATGGGTTAGGTGTAACTCGTGAAGTAGGCCAGGACTTAGTAAATTTAAATGCTAAAAATGTATGCGTCATGACAGACCCTAATGTTGCATCTTTGAGTCCTATGAAAGCTGTTCTGGATTCACTCACTAGAAATGGAATAAATTATAAAGTTTATGATAAAGTGAGAGTAGAACCTACAGATGGCAGTTTCAAAGATGCAATTCATTTTGCAAAGGAAGGAAACTTTGACAGTTTTGTTGCTATTGGTGGGGGATCAGTGATGGATACTTGTAAAGCTGCCAATTTGTACTATAGTGACCCTGATGCTGAGTTCTTGGATTATGTGAATGCTCCTGTTGGCAAAGGCAAAGCTGTCAATATACAGGTTAAACCAATGATTGCCATACCAACTACGAGCGGCACGGGCAGCGAGACCACCGGCGTCAGCATTTTCGATTACGAAGAAATCCACGCTAAAACCGGTATATCTCATAATGCTCTCCGACCGATACTAGCGTTAATTGATCCGCTTCACACTTTGACAGTGCCCGAGAAAGTATCAAATTATTCTGGTTTCGATATATTTTGCCACGCTTTGGAAAGCTTCACAGCGATACCGTTCACTGAAAGAGGGCCCGCGCCTGCTAATCCGGCTCTCAGACCAGCATATCAAGGCAGCAATCCCATATCTGATGTTTGGGCGCGATTTTGTTTGCAGACCCTGCAAAAGTATTTCGTTCGTTCAGTGAACAATTCTGGAGACGTGGAAGCGAGATCGAGCATGCATCTGGCGGCGACGATGGCGGGCGTCGGCATCGGAAACGCGGGCGTACATTTATGTCACGGGCTAGCGTACCCGATAGCTGGAAACGTTAAGAGTTTTGTTCCCGAGGACTATGGTTCCAATCCGATAATCCCTCACGGCCTGTCGGTTGTAATGACAGCGCCGGCTGTTTTCCGATTCACCGCATCTAGTGACCCTGAGAAACATCTAGAAGCAGCCAGTCTGCTAGGAGCTGACGTCACCAACGCTAAGAGAAAAGACGCCGGTAGAATACTCTCCGACGTCATACTCTTGTATATGGATAAGTTGAAGATTGAAAACGGATTGAAGGCATTAGGATATACTAATGACGACATACCGGACTTGGTCAAGGGTGCTTTACCTCAGGATCGTTTATTAAAAATCGCTCCAGTGACACAATCTGAAGAGGATTTATCAAAATTGCTGGAGGAATCATTGACCATTTACTAA
Protein
MTSRKRIFDLLRTINIASCQCPAHGYRGNIQVSNATPLKDYAFEIKCSTVRYGLGVTREVGQDLVNLNAKNVCVMTDPNVASLSPMKAVLDSLTRNGINYKVYDKVRVEPTDGSFKDAIHFAKEGNFDSFVAIGGGSVMDTCKAANLYYSDPDAEFLDYVNAPVGKGKAVNIQVKPMIAIPTTSGTGSETTGVSIFDYEEIHAKTGISHNALRPILALIDPLHTLTVPEKVSNYSGFDIFCHALESFTAIPFTERGPAPANPALRPAYQGSNPISDVWARFCLQTLQKYFVRSVNNSGDVEARSSMHLAATMAGVGIGNAGVHLCHGLAYPIAGNVKSFVPEDYGSNPIIPHGLSVVMTAPAVFRFTASSDPEKHLEAASLLGADVTNAKRKDAGRILSDVILLYMDKLKIENGLKALGYTNDDIPDLVKGALPQDRLLKIAPVTQSEEDLSKLLEESLTIY
Summary
Description
Catalyzes the cofactor-independent reversible oxidation of gamma-hydroxybutyrate (GHB) to succinic semialdehyde (SSA) coupled to reduction of 2-ketoglutarate (2-KG) to D-2-hydroxyglutarate (D-2-HG). L-3-hydroxybutyrate (L-3-OHB) is also a substrate for HOT when using 2-KG as hydrogen acceptor, resulting in the formation of D-2-HG (By similarity).
Catalytic Activity
(S)-3-hydroxybutanoate + 2-oxoglutarate = (R)-2-hydroxyglutarate + acetoacetate
2-oxoglutarate + 4-hydroxybutanoate = (R)-2-hydroxyglutarate + succinate semialdehyde
2-oxoglutarate + 4-hydroxybutanoate = (R)-2-hydroxyglutarate + succinate semialdehyde
Similarity
Belongs to the iron-containing alcohol dehydrogenase family. Hydroxyacid-oxoacid transhydrogenase subfamily.
Belongs to the glyceraldehyde-3-phosphate dehydrogenase family.
Belongs to the glyceraldehyde-3-phosphate dehydrogenase family.
Keywords
Complete proteome
Mitochondrion
Oxidoreductase
Reference proteome
Transit peptide
Feature
chain Probable hydroxyacid-oxoacid transhydrogenase, mitochondrial
Uniprot
A0A2H1WB69
A0A2A4J8N0
A0A212FLL8
A0A194Q2C9
A0A067QTV8
A0A2J7RB98
+ More
E2ABQ4 A0A146KTV5 A0A0A9XR32 D6W8A2 A0A0V0G9S6 E9INY1 A0A026WZP0 A0A2M4ANA6 A0A2R7X5F0 A0A069DTV5 A0A1S3D6P1 F4WH16 A0A1B6M259 A0A182JI21 A0A0P4W3B5 R4FQE2 Q17EN4 A0A182RDJ8 W5JCR3 A0A182FDB3 A0A2K7P695 A0A158NWM4 A0A182WBM3 A0A182M6J0 A0A023F7W9 A0A182QFM5 A0A158NWM5 A0A182NI63 A0A2S2PZR8 A0A151X5L4 A0A195BSZ1 A0A182PDJ5 A0A1B6CDA2 A0A3F2Z247 A0A151K0T9 A0A2S2NN16 A0A182SFB8 Q17EN5 A0A182I4M4 A0A182Y2G2 A0A182V943 Q7Q547 B0WEG0 A0A2A3E6J9 J9KA60 A0A0P4W8T3 A0A1B0CPT7 A0A1B6G2G3 A0A182U1C9 A0A2H8TU81 A0A2L2Y052 E2BXU4 V9KER4 A0A195C8W3 A0A087TZ15 A0A084WMD3 A0A194R770 U5EWU8 A0A154PAN7 A0A1S4F5U5 B4I831 W5MBF6 A0A1S3LXT4 A0A060X9F1 A0A195E351 B3NNM9 A0A1L8E0J8 A0A3P9MJ96 A0A2M4CMK4 A0A3P9I8T3 A0A158NT98 A0A3P8ZT68 H2LR58 G3N9N5 A0A3P9CVV0 B4P7W3 A0A023GNG8 B4QH42 A0A3B3Y1T2 W8BE93 A0A158NT99 A0A3B3TDE4 A0A0L7R4C6 A0A3B0J5U8 A0A146XFT9 A0A1W5ABE9 S5YSM1 I3IVJ4 A0A146XEM5 A0A034WGM7 A0A3B3CP14 B4GHJ1 B4MJH2 Q28XT3
E2ABQ4 A0A146KTV5 A0A0A9XR32 D6W8A2 A0A0V0G9S6 E9INY1 A0A026WZP0 A0A2M4ANA6 A0A2R7X5F0 A0A069DTV5 A0A1S3D6P1 F4WH16 A0A1B6M259 A0A182JI21 A0A0P4W3B5 R4FQE2 Q17EN4 A0A182RDJ8 W5JCR3 A0A182FDB3 A0A2K7P695 A0A158NWM4 A0A182WBM3 A0A182M6J0 A0A023F7W9 A0A182QFM5 A0A158NWM5 A0A182NI63 A0A2S2PZR8 A0A151X5L4 A0A195BSZ1 A0A182PDJ5 A0A1B6CDA2 A0A3F2Z247 A0A151K0T9 A0A2S2NN16 A0A182SFB8 Q17EN5 A0A182I4M4 A0A182Y2G2 A0A182V943 Q7Q547 B0WEG0 A0A2A3E6J9 J9KA60 A0A0P4W8T3 A0A1B0CPT7 A0A1B6G2G3 A0A182U1C9 A0A2H8TU81 A0A2L2Y052 E2BXU4 V9KER4 A0A195C8W3 A0A087TZ15 A0A084WMD3 A0A194R770 U5EWU8 A0A154PAN7 A0A1S4F5U5 B4I831 W5MBF6 A0A1S3LXT4 A0A060X9F1 A0A195E351 B3NNM9 A0A1L8E0J8 A0A3P9MJ96 A0A2M4CMK4 A0A3P9I8T3 A0A158NT98 A0A3P8ZT68 H2LR58 G3N9N5 A0A3P9CVV0 B4P7W3 A0A023GNG8 B4QH42 A0A3B3Y1T2 W8BE93 A0A158NT99 A0A3B3TDE4 A0A0L7R4C6 A0A3B0J5U8 A0A146XFT9 A0A1W5ABE9 S5YSM1 I3IVJ4 A0A146XEM5 A0A034WGM7 A0A3B3CP14 B4GHJ1 B4MJH2 Q28XT3
EC Number
1.1.99.24
Pubmed
22118469
26354079
24845553
20798317
26823975
25401762
+ More
18362917 19820115 21282665 24508170 30249741 26334808 21719571 27129103 17510324 20920257 23761445 21347285 25474469 25244985 12364791 26561354 24402279 24438588 17994087 24755649 17554307 25069045 25186727 17550304 22936249 24495485 29240929 25348373 29451363 15632085
18362917 19820115 21282665 24508170 30249741 26334808 21719571 27129103 17510324 20920257 23761445 21347285 25474469 25244985 12364791 26561354 24402279 24438588 17994087 24755649 17554307 25069045 25186727 17550304 22936249 24495485 29240929 25348373 29451363 15632085
EMBL
ODYU01007382
SOQ50092.1
NWSH01002374
PCG68435.1
AGBW02007743
OWR54631.1
+ More
KQ459589 KPI97560.1 KK852945 KDR13466.1 NEVH01005921 PNF38105.1 GL438334 EFN69156.1 GDHC01020037 JAP98591.1 GBHO01020437 GBHO01011259 GBRD01008098 GBRD01007315 JAG23167.1 JAG32345.1 JAG57723.1 KQ971307 EFA10918.1 GECL01001225 JAP04899.1 GL764397 EFZ17696.1 KK107061 QOIP01000014 EZA61308.1 RLU15004.1 GGFK01008943 MBW42264.1 KK856984 PTY26650.1 GBGD01001366 JAC87523.1 GL888148 EGI66495.1 GEBQ01009956 JAT30021.1 GDKW01000646 JAI55949.1 ACPB03009797 GAHY01000444 JAA77066.1 CH477280 ADMH02001892 ETN60619.1 ADTU01028207 AXCM01002363 GBBI01001365 JAC17347.1 AXCN02000396 GGMS01001804 MBY71007.1 KQ982515 KYQ55614.1 KQ976409 KYM90944.1 GEDC01025896 JAS11402.1 KQ981248 KYN44264.1 GGMR01005925 MBY18544.1 EAT44959.1 APCN01002331 AAAB01008960 DS231909 EDS45620.1 KZ288365 PBC26846.1 ABLF02039218 GDRN01081512 JAI62006.1 AJWK01022638 GECZ01017703 GECZ01017586 GECZ01017581 GECZ01016103 GECZ01013142 JAS52066.1 JAS52183.1 JAS52188.1 JAS53666.1 JAS56627.1 GFXV01005695 MBW17500.1 IAAA01006077 IAAA01006078 LAA00485.1 GL451314 EFN79501.1 JW863944 AFO96461.1 KQ978072 KYM97267.1 KK117391 KFM70354.1 ATLV01024425 KE525352 KFB51377.1 KQ460870 KPJ11716.1 GANO01001259 JAB58612.1 KQ434860 KZC08913.1 CH480824 EDW56756.1 AHAT01004377 FR904940 CDQ73949.1 KQ979701 KYN19595.1 CH954179 EDV55586.1 GFDF01001836 JAV12248.1 GGFL01002394 MBW66572.1 ADTU01025758 CM000158 EDW92118.1 GBBM01001008 JAC34410.1 CM000362 CM002911 EDX08188.1 KMY95766.1 GAMC01006945 JAB99610.1 KQ414659 KOC65611.1 OUUW01000001 SPP75082.1 GCES01045799 JAR40524.1 KF439724 AGT15702.1 AERX01045149 GCES01045800 JAR40523.1 GAKP01006024 JAC52928.1 CH479183 EDW35961.1 CH963846 EDW72261.1 CM000071
KQ459589 KPI97560.1 KK852945 KDR13466.1 NEVH01005921 PNF38105.1 GL438334 EFN69156.1 GDHC01020037 JAP98591.1 GBHO01020437 GBHO01011259 GBRD01008098 GBRD01007315 JAG23167.1 JAG32345.1 JAG57723.1 KQ971307 EFA10918.1 GECL01001225 JAP04899.1 GL764397 EFZ17696.1 KK107061 QOIP01000014 EZA61308.1 RLU15004.1 GGFK01008943 MBW42264.1 KK856984 PTY26650.1 GBGD01001366 JAC87523.1 GL888148 EGI66495.1 GEBQ01009956 JAT30021.1 GDKW01000646 JAI55949.1 ACPB03009797 GAHY01000444 JAA77066.1 CH477280 ADMH02001892 ETN60619.1 ADTU01028207 AXCM01002363 GBBI01001365 JAC17347.1 AXCN02000396 GGMS01001804 MBY71007.1 KQ982515 KYQ55614.1 KQ976409 KYM90944.1 GEDC01025896 JAS11402.1 KQ981248 KYN44264.1 GGMR01005925 MBY18544.1 EAT44959.1 APCN01002331 AAAB01008960 DS231909 EDS45620.1 KZ288365 PBC26846.1 ABLF02039218 GDRN01081512 JAI62006.1 AJWK01022638 GECZ01017703 GECZ01017586 GECZ01017581 GECZ01016103 GECZ01013142 JAS52066.1 JAS52183.1 JAS52188.1 JAS53666.1 JAS56627.1 GFXV01005695 MBW17500.1 IAAA01006077 IAAA01006078 LAA00485.1 GL451314 EFN79501.1 JW863944 AFO96461.1 KQ978072 KYM97267.1 KK117391 KFM70354.1 ATLV01024425 KE525352 KFB51377.1 KQ460870 KPJ11716.1 GANO01001259 JAB58612.1 KQ434860 KZC08913.1 CH480824 EDW56756.1 AHAT01004377 FR904940 CDQ73949.1 KQ979701 KYN19595.1 CH954179 EDV55586.1 GFDF01001836 JAV12248.1 GGFL01002394 MBW66572.1 ADTU01025758 CM000158 EDW92118.1 GBBM01001008 JAC34410.1 CM000362 CM002911 EDX08188.1 KMY95766.1 GAMC01006945 JAB99610.1 KQ414659 KOC65611.1 OUUW01000001 SPP75082.1 GCES01045799 JAR40524.1 KF439724 AGT15702.1 AERX01045149 GCES01045800 JAR40523.1 GAKP01006024 JAC52928.1 CH479183 EDW35961.1 CH963846 EDW72261.1 CM000071
Proteomes
UP000218220
UP000007151
UP000053268
UP000027135
UP000235965
UP000000311
+ More
UP000007266 UP000053097 UP000279307 UP000079169 UP000007755 UP000075880 UP000015103 UP000008820 UP000075900 UP000000673 UP000069272 UP000005205 UP000075920 UP000075883 UP000075886 UP000075884 UP000075809 UP000078540 UP000075885 UP000076407 UP000078541 UP000075901 UP000075840 UP000076408 UP000075903 UP000007062 UP000002320 UP000242457 UP000007819 UP000092461 UP000075902 UP000008237 UP000078542 UP000054359 UP000030765 UP000053240 UP000076502 UP000001292 UP000018468 UP000087266 UP000193380 UP000078492 UP000008711 UP000265180 UP000265200 UP000265140 UP000001038 UP000007635 UP000265160 UP000002282 UP000000304 UP000261480 UP000261540 UP000053825 UP000268350 UP000192224 UP000005207 UP000261560 UP000008744 UP000007798 UP000001819
UP000007266 UP000053097 UP000279307 UP000079169 UP000007755 UP000075880 UP000015103 UP000008820 UP000075900 UP000000673 UP000069272 UP000005205 UP000075920 UP000075883 UP000075886 UP000075884 UP000075809 UP000078540 UP000075885 UP000076407 UP000078541 UP000075901 UP000075840 UP000076408 UP000075903 UP000007062 UP000002320 UP000242457 UP000007819 UP000092461 UP000075902 UP000008237 UP000078542 UP000054359 UP000030765 UP000053240 UP000076502 UP000001292 UP000018468 UP000087266 UP000193380 UP000078492 UP000008711 UP000265180 UP000265200 UP000265140 UP000001038 UP000007635 UP000265160 UP000002282 UP000000304 UP000261480 UP000261540 UP000053825 UP000268350 UP000192224 UP000005207 UP000261560 UP000008744 UP000007798 UP000001819
PRIDE
Interpro
IPR039697
Alcohol_dehydrogenase_Fe
+ More
IPR001670 ADH_Fe/GldA
IPR042157 HOT
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR020828 GlycerAld_3-P_DH_NAD(P)-bd
IPR020831 GlycerAld/Erythrose_P_DH
IPR020829 GlycerAld_3-P_DH_cat
IPR036291 NAD(P)-bd_dom_sf
IPR020830 GlycerAld_3-P_DH_AS
IPR006424 Glyceraldehyde-3-P_DH_1
IPR001670 ADH_Fe/GldA
IPR042157 HOT
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR020828 GlycerAld_3-P_DH_NAD(P)-bd
IPR020831 GlycerAld/Erythrose_P_DH
IPR020829 GlycerAld_3-P_DH_cat
IPR036291 NAD(P)-bd_dom_sf
IPR020830 GlycerAld_3-P_DH_AS
IPR006424 Glyceraldehyde-3-P_DH_1
SUPFAM
SSF51735
SSF51735
Gene 3D
CDD
ProteinModelPortal
A0A2H1WB69
A0A2A4J8N0
A0A212FLL8
A0A194Q2C9
A0A067QTV8
A0A2J7RB98
+ More
E2ABQ4 A0A146KTV5 A0A0A9XR32 D6W8A2 A0A0V0G9S6 E9INY1 A0A026WZP0 A0A2M4ANA6 A0A2R7X5F0 A0A069DTV5 A0A1S3D6P1 F4WH16 A0A1B6M259 A0A182JI21 A0A0P4W3B5 R4FQE2 Q17EN4 A0A182RDJ8 W5JCR3 A0A182FDB3 A0A2K7P695 A0A158NWM4 A0A182WBM3 A0A182M6J0 A0A023F7W9 A0A182QFM5 A0A158NWM5 A0A182NI63 A0A2S2PZR8 A0A151X5L4 A0A195BSZ1 A0A182PDJ5 A0A1B6CDA2 A0A3F2Z247 A0A151K0T9 A0A2S2NN16 A0A182SFB8 Q17EN5 A0A182I4M4 A0A182Y2G2 A0A182V943 Q7Q547 B0WEG0 A0A2A3E6J9 J9KA60 A0A0P4W8T3 A0A1B0CPT7 A0A1B6G2G3 A0A182U1C9 A0A2H8TU81 A0A2L2Y052 E2BXU4 V9KER4 A0A195C8W3 A0A087TZ15 A0A084WMD3 A0A194R770 U5EWU8 A0A154PAN7 A0A1S4F5U5 B4I831 W5MBF6 A0A1S3LXT4 A0A060X9F1 A0A195E351 B3NNM9 A0A1L8E0J8 A0A3P9MJ96 A0A2M4CMK4 A0A3P9I8T3 A0A158NT98 A0A3P8ZT68 H2LR58 G3N9N5 A0A3P9CVV0 B4P7W3 A0A023GNG8 B4QH42 A0A3B3Y1T2 W8BE93 A0A158NT99 A0A3B3TDE4 A0A0L7R4C6 A0A3B0J5U8 A0A146XFT9 A0A1W5ABE9 S5YSM1 I3IVJ4 A0A146XEM5 A0A034WGM7 A0A3B3CP14 B4GHJ1 B4MJH2 Q28XT3
E2ABQ4 A0A146KTV5 A0A0A9XR32 D6W8A2 A0A0V0G9S6 E9INY1 A0A026WZP0 A0A2M4ANA6 A0A2R7X5F0 A0A069DTV5 A0A1S3D6P1 F4WH16 A0A1B6M259 A0A182JI21 A0A0P4W3B5 R4FQE2 Q17EN4 A0A182RDJ8 W5JCR3 A0A182FDB3 A0A2K7P695 A0A158NWM4 A0A182WBM3 A0A182M6J0 A0A023F7W9 A0A182QFM5 A0A158NWM5 A0A182NI63 A0A2S2PZR8 A0A151X5L4 A0A195BSZ1 A0A182PDJ5 A0A1B6CDA2 A0A3F2Z247 A0A151K0T9 A0A2S2NN16 A0A182SFB8 Q17EN5 A0A182I4M4 A0A182Y2G2 A0A182V943 Q7Q547 B0WEG0 A0A2A3E6J9 J9KA60 A0A0P4W8T3 A0A1B0CPT7 A0A1B6G2G3 A0A182U1C9 A0A2H8TU81 A0A2L2Y052 E2BXU4 V9KER4 A0A195C8W3 A0A087TZ15 A0A084WMD3 A0A194R770 U5EWU8 A0A154PAN7 A0A1S4F5U5 B4I831 W5MBF6 A0A1S3LXT4 A0A060X9F1 A0A195E351 B3NNM9 A0A1L8E0J8 A0A3P9MJ96 A0A2M4CMK4 A0A3P9I8T3 A0A158NT98 A0A3P8ZT68 H2LR58 G3N9N5 A0A3P9CVV0 B4P7W3 A0A023GNG8 B4QH42 A0A3B3Y1T2 W8BE93 A0A158NT99 A0A3B3TDE4 A0A0L7R4C6 A0A3B0J5U8 A0A146XFT9 A0A1W5ABE9 S5YSM1 I3IVJ4 A0A146XEM5 A0A034WGM7 A0A3B3CP14 B4GHJ1 B4MJH2 Q28XT3
PDB
4FR2
E-value=1.30999e-22,
Score=264
Ontologies
GO
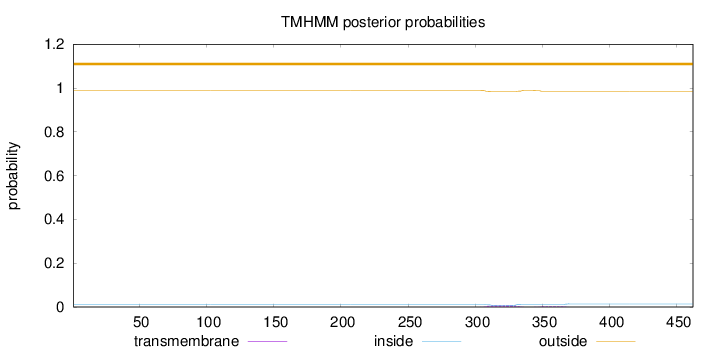
Topology
Subcellular location
Mitochondrion
Length:
462
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25724
Exp number, first 60 AAs:
0.00076
Total prob of N-in:
0.01155
outside
1 - 462
Population Genetic Test Statistics
Pi
238.466773
Theta
198.290298
Tajima's D
0.643332
CLR
0
CSRT
0.554322283885806
Interpretation
Uncertain
