Pre Gene Modal
BGIBMGA002618
Annotation
PREDICTED:_uncharacterized_protein_LOC106124291_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.325
Sequence
CDS
ATGGGGGCAAGGTCCCGGGAGTTACTACTGCTCCTGGCAGCATGCGCGCTGGTATTCGGCGAATCTATCAATAAAACCGTCAAGAAAGACAAAGACGCCGACAACTTATTGGATCAATATGAGGATTACGAACCCGCGGAATACCAAGAGGTTTTGTACAACGAGGACAGGCCCTGTCCCAGAGACTGCATATGCTCTGTATCTCAGGGATACAGACAAGCCAAGTGCAGCTTCCTCGAAATCGGTACACAAAAGTTCGGCGATGATATCCTTGATCTCGTGGTCGAAAACGCAGATCCCCGCTACCCGATTAATCTAGACGACTTCATGTTCAAAAAACTCGGACTTCACCAGGTTGCTACTGTAAAAATCGTGAACAGCACGATTGGTTATATCGCCCCCAATGCTTTCCATGGTGTCCATGACCTTTACGCAGTCAACCTATCCAACAACAACCTAAAGAGCCTTCATCCTGAGACGTTTGCGAACAATAAGAAGCTGTTACTTCTAACGCTTTCAAACAATCCCCTCAAATTTCCTGCAGCTGGGTCCGAAAATTACTTCTTGAACGCTTCCTCTGTACAAGAGCTTGATGTTTCATACTGTAACATGCAGTACATCACGGCAAATACTTTCAAAAATATGCCTGGTTTGATGTACTTGAATGTTGCCGGTAACAACCTATCCGATATGGATCCAGATACATTCAAAAAATTACTCGATCTCGAAGAGTTAGATCTCCGCAACAATCACATTAAATCGCTCCCCGACGACATTTTCTCGGAAAACACCGAACTAGCAACACTGCATTTACTGAAAAATCCTATTGATACAGTGTATGGCCTCCAAATATCCGATTTGCTTACACTTAATGCGGGACAAACAAACATTAAGTTTGTTGGGCCATCAATGTTCAACGGTATGGGTCTTATTGCTAATCTCAACCTCAGCGGAAATAGTATCGAGAAAATCCATAACCAGGCATTCCACAAATTAGTAGAACTCAATTATTTAGATTTGTCGTATAATAATTTGGATTACATGTCCAGTATTCTTATCAAAGAAAACATCGAACTTGACATCTTCAAAATTTCAAACAACCCACGGCTAAAACATTTACCGGTTGAAGGTTTCACATGCTCATCTGATCAGTTTAATATTTACTTCTTTGGCGCATCTGACTGTGGTCTAGAAGAGATTTACGATAATTCTCTGAAAACATTCTCTGCTCTGTCAATAATTCATCTATCGAATAATAAATTAACTACCATAAACAATACCGTTTTCTCGCACTGCCCTAAATTAATGGAAATCAATCTTGCTTATAATATGCTCACCACTTTAGATCCTAAGCTCTTCGCAAAGAACAATGAACTTGCAAAGCTTAATTTACAAGGTAACCCTCTTAAAGTACTTTCTGCGGAAATTTTTGTCCACACTCCGGCCATGATTTGGCTAGACATGAGTCACGCTGAATTAACCTCTTTGTGGAAAGTAGATAAAAATCACCCACTGACAGTTCTTATCAATTTACTATTCTTGAATGTATCCCACAATAGAATTACGGAAATTAAACAAAACGAATTAGATAATTTGAACAAACTACGTACTTTAGACATTACCAACAATCCGTTGGCCTGCAGCCGAGACTTTGAAAACCTCATGACATGGCTGAACAACCACACAGTGTTGCCTACTGGTAACTCTGCCAACGTCGCCAATATGGCTAGAGATGGAAAAGATGATGATGCCACTTACAGCTGGGATTTCCTGACTATCAAGACTTGTGGAACTACAGTTATTTCCCATCCTGTTGAACCATTATCGGCTGTATCTGACGAAGAAATTTGGGAAAGAATCGACAAGGACACTGAGGGTAATTTCGATTTGAAGAACACCTTAGATGACGGTAAAATTCCTGATGATACCAACGCTTCTGCTGACAGCAAACTAATAATTGATAGCACTGATGAAGAATCAGATGACGTTGATCCCGATGATGATGAAGACGAGGGCGACGATGATGACGACGAAGACGACTCTGGCGAAGATTACGAGAATGATGACATAGATCTTAAATATGAGCTCATCAACAAAACGGACTCGAAATCTGAGAATACAAAGAAATCTGAACAACAGGCCACAGAGAAAGTTAAGATTGACATCAAGCTCTTAGACTTATACGAAAATTCTGAGCCAGACGTCTATGTACGAACATCGACCGAGGAAGAACACGGCCATTACAGCTACTTGTGGCCAATATCCATTGCCATCCTCGGAGCGCTTTTGTTGCTAATCGTTATCGCTAAGGTCGTCATGATGACGTGTAAATCAAGAACCAGGCAGCTCAGATACAACAGTGCCATTATTGCAGCGATGAGTCAACCCGGCCGCACCAAAAAGGACTGCGGCTTGGTATACCAGCAGTTGTCCGAAGACCTCACAGGTCCGGCCACTCCAAAACTAAGCCGATATGCTCCACTTCACAGCGTCACTGTCAATGCATCCAACATGTCCTATGAGAGCAGTCCCTTCCACCACAACAACATCGTGCCAGAAGCAGTGTGA
Protein
MGARSRELLLLLAACALVFGESINKTVKKDKDADNLLDQYEDYEPAEYQEVLYNEDRPCPRDCICSVSQGYRQAKCSFLEIGTQKFGDDILDLVVENADPRYPINLDDFMFKKLGLHQVATVKIVNSTIGYIAPNAFHGVHDLYAVNLSNNNLKSLHPETFANNKKLLLLTLSNNPLKFPAAGSENYFLNASSVQELDVSYCNMQYITANTFKNMPGLMYLNVAGNNLSDMDPDTFKKLLDLEELDLRNNHIKSLPDDIFSENTELATLHLLKNPIDTVYGLQISDLLTLNAGQTNIKFVGPSMFNGMGLIANLNLSGNSIEKIHNQAFHKLVELNYLDLSYNNLDYMSSILIKENIELDIFKISNNPRLKHLPVEGFTCSSDQFNIYFFGASDCGLEEIYDNSLKTFSALSIIHLSNNKLTTINNTVFSHCPKLMEINLAYNMLTTLDPKLFAKNNELAKLNLQGNPLKVLSAEIFVHTPAMIWLDMSHAELTSLWKVDKNHPLTVLINLLFLNVSHNRITEIKQNELDNLNKLRTLDITNNPLACSRDFENLMTWLNNHTVLPTGNSANVANMARDGKDDDATYSWDFLTIKTCGTTVISHPVEPLSAVSDEEIWERIDKDTEGNFDLKNTLDDGKIPDDTNASADSKLIIDSTDEESDDVDPDDDEDEGDDDDDEDDSGEDYENDDIDLKYELINKTDSKSENTKKSEQQATEKVKIDIKLLDLYENSEPDVYVRTSTEEEHGHYSYLWPISIAILGALLLLIVIAKVVMMTCKSRTRQLRYNSAIIAAMSQPGRTKKDCGLVYQQLSEDLTGPATPKLSRYAPLHSVTVNASNMSYESSPFHHNNIVPEAV
Summary
Uniprot
EMBL
BABH01014261
BABH01014262
KQ459589
KPI97561.1
AGBW02007743
OWR54632.1
+ More
ODYU01007382 SOQ50093.1 NWSH01002374 PCG68439.1 GAIX01005179 JAA87381.1 GDQN01006535 GDQN01003121 JAT84519.1 JAT87933.1 JTDY01004694 KOB67866.1 MG846897 AXY94749.1 OUUW01000001 SPP75083.1 JRES01001423 KNC23005.1 CH479183 EDW35960.1 CM000071 EAL26234.2 AF151684 AAF67245.1
ODYU01007382 SOQ50093.1 NWSH01002374 PCG68439.1 GAIX01005179 JAA87381.1 GDQN01006535 GDQN01003121 JAT84519.1 JAT87933.1 JTDY01004694 KOB67866.1 MG846897 AXY94749.1 OUUW01000001 SPP75083.1 JRES01001423 KNC23005.1 CH479183 EDW35960.1 CM000071 EAL26234.2 AF151684 AAF67245.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
4LXS
E-value=1.0843e-13,
Score=189
Ontologies
KEGG
GO
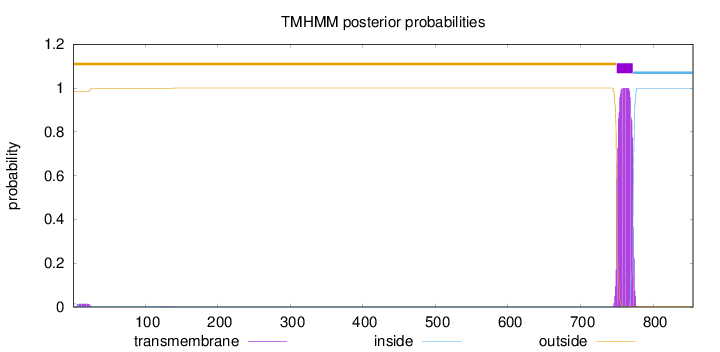
Topology
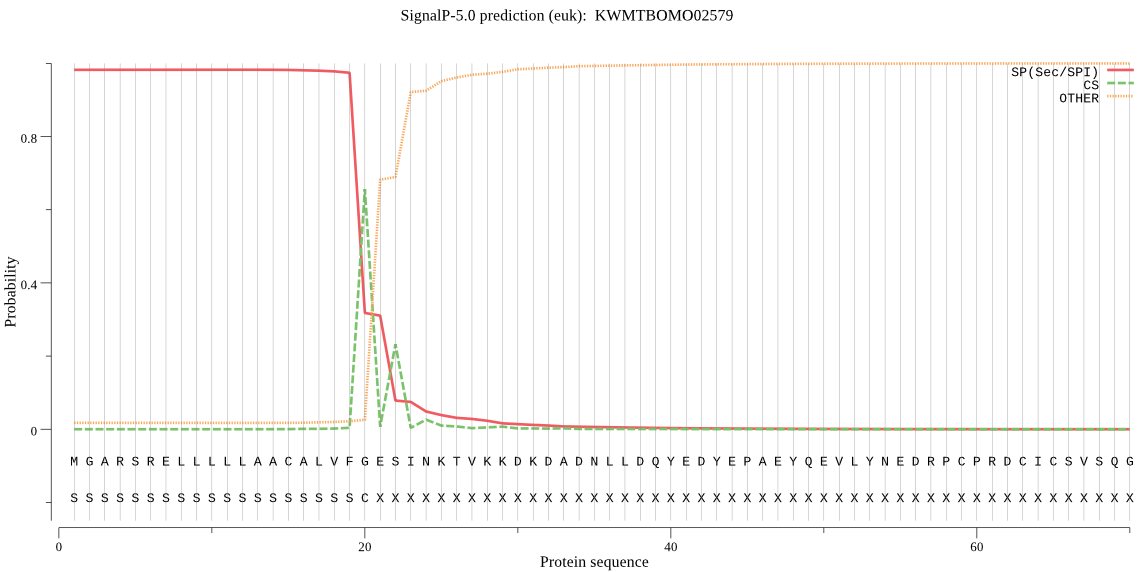
SignalP
Position: 1 - 20,
Likelihood: 0.982459
Length:
855
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.72632
Exp number, first 60 AAs:
0.25013
Total prob of N-in:
0.01446
outside
1 - 749
TMhelix
750 - 772
inside
773 - 855
Population Genetic Test Statistics
Pi
277.716852
Theta
36.534405
Tajima's D
0.583846
CLR
0.736886
CSRT
0.540972951352432
Interpretation
Uncertain
