Gene
KWMTBOMO02576
Pre Gene Modal
BGIBMGA002617
Annotation
Leucine-rich_transmembrane_protein_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.951
Sequence
CDS
ATGTTACGACTGTCCAGCGTATGGATTATTCTGATATCATGGAATTTGTTAGCGCTTGCAAGAAGCTTGGATGACGATGACAAAACCATAGGAGATTTGTGTTTTCTATGCAGATGTAATCTAGATAAAACTGATATTGATTGTTCACGTAGAGGCCTCACGGACATCCCCAATGGATTGGGATTACAGGTGACCAAACTGAACATTTCGAATAATGAGTTTACAAAATTTCCCGAGTCTCTGAGCAGACTCCACAACCTCGTGAATTTGGATATTAGCAGCAACCAATTGAAGGGATTGCCTGACAATGCGCTTTACAATTTAACAGCGCTCGAAGTGCTGAATTTATCAAGAAATTACTTTGATTCGTGGCTTAATTTGAATCCGAACGATGTACTGCTCCCGGCTACACACTTGAAAATTCTGGACCTGTCGTTTAACAAATTTCAGACTATGGGCAATTTGGCCAATCAAGAATTATTGATCAGCCCATCCCTGGAAACGTTAATATTAGATCATTGTGAAATAAATTCCATACATGGTAGGTCTCCATTAAGTGGTCTGATAAACATAAGAGTTCTAAAGTTGAACTTCAATCCTTTATTGAGAATACAAAATTTAATATCGTCATCCCTGAGGAGTCTTTTCGTTAGTAACTGTGAGCTGACCTCATTAAATCATAACGAGTTGATGTTCTTACCTTCGTTGACTCATTTAAAAATGTCAAATAACTATCGTCTCGAGTTAGCGTCTTCAGCGAACAATTTGTTTTCGCAATCATTGAAATATATCGATATTTCATACTGTAATATTTTGCAACCGAACCTCAAGGGGTTCCCTAGTTTGCGAAAAGCTATCCTAAATCACAATATGGTTAGGTATTTAGAAAGTAACGCTTTTGCTAACAATACAGAATTGGAATATTTGGATTTGTCCAATAACAACATTGCGTCGCTTAGATACGATACGTTTCGAGGACTGAAGATGCTGAAACACTTGGATTTATCTTGGAACGAAATCGCTGTGATTCCAGAAGATAGCTTATTGGAAATGCCGTCTTTGACGCAATTGAAATTATCCAGAAATTACCTGTCGAGAGTTGGTCACTTGAGATCAATGTCTGTAACGATTCTCGATATGAGTTCATGTGAATTAAATACAATCGGTAAAGATTCACTGGAGGGCTTGCAGTCGATTGTTGAATTAGATTTGTCTCAGAATCTGCTGTCTTATATTCCGGATAGTATTTCTTCGAACACTTTGAAATACCTTAATTTGAATTATAATCGAATTAGCAGCATCAATAATTTAACGTTTTTCATGTTGAATCGGTTGACTAGCTTAAGCGTGGTCGGTAACAGATTCACTATGATCTGGCGGAGATCGTTTTTCGATTCCAATCCATATTTGGAAAGACTGGATTTGAGTGACAACATGTGGAGATGTGATTGCTCCGATGAGAACATGAGGGACTTTTACGACTTTGTCACTTTAGAACCAAATAAAAAAGAAGAATCGTTCAATCTTATATGCAATAGTCCGGCTAATGTGAACACGCAGACGTGGCTCGAGGCGTGTTACTTTACTTGGAATCCCACCGAAAAAACCGCTAACGCAGACACTTTAATATGGTTTATTGTTGTCATGATCATAGGCTTGTCCTTGTGTATATTATTAGTGAGCGGAATCCGAAGTTCGATGAAACGTCGTCTTGCCGCCATACAAGCAGAAAGGGAAAGGCAAGTGGAAGAAGCTAGAGACAGATTGAGGCAAATGAGAATACGTGCGGAACAGGAAGCGTTATGTAATACACCAGATCCACGTGATTTGATCGCGCCACCGTCGTACGATGAAGCTCTCTCGATGCCAAAACTAAATGTATCGTGTCACTCTTTGAACGAAACCGGTTCAGGAAAAACCAGGAGAAAGAGAGGGAGAAGGAAAACGAAATCGAGTGGTGATCTATTAGAAGAAACTGACAGGAACGGTGATGCACCCGTATATGATGATTTAGAACTAACCGAGACCCAAGAGGAAGATAGACAGCGGAGGCGTCGTAGAAGAAATAGAAGGTATGGCAGTCACGAAATAGCTGAGTTGGATCAATCACCAGGTGCTCGGCGTCGGCGAATGTCTGAATATATTGTTGAAAACGATGATAGTGTTATTGTGGAAGTTCAAGCGCAGTTAGAACGACCCCTTCGCCCTAGAAATCGACGGTATTCGATTGATGATAATGATGTCAGGGAAAGTGACTTTTAA
Protein
MLRLSSVWIILISWNLLALARSLDDDDKTIGDLCFLCRCNLDKTDIDCSRRGLTDIPNGLGLQVTKLNISNNEFTKFPESLSRLHNLVNLDISSNQLKGLPDNALYNLTALEVLNLSRNYFDSWLNLNPNDVLLPATHLKILDLSFNKFQTMGNLANQELLISPSLETLILDHCEINSIHGRSPLSGLINIRVLKLNFNPLLRIQNLISSSLRSLFVSNCELTSLNHNELMFLPSLTHLKMSNNYRLELASSANNLFSQSLKYIDISYCNILQPNLKGFPSLRKAILNHNMVRYLESNAFANNTELEYLDLSNNNIASLRYDTFRGLKMLKHLDLSWNEIAVIPEDSLLEMPSLTQLKLSRNYLSRVGHLRSMSVTILDMSSCELNTIGKDSLEGLQSIVELDLSQNLLSYIPDSISSNTLKYLNLNYNRISSINNLTFFMLNRLTSLSVVGNRFTMIWRRSFFDSNPYLERLDLSDNMWRCDCSDENMRDFYDFVTLEPNKKEESFNLICNSPANVNTQTWLEACYFTWNPTEKTANADTLIWFIVVMIIGLSLCILLVSGIRSSMKRRLAAIQAERERQVEEARDRLRQMRIRAEQEALCNTPDPRDLIAPPSYDEALSMPKLNVSCHSLNETGSGKTRRKRGRRKTKSSGDLLEETDRNGDAPVYDDLELTETQEEDRQRRRRRRNRRYGSHEIAELDQSPGARRRRMSEYIVENDDSVIVEVQAQLERPLRPRNRRYSIDDNDVRESDF
Summary
Uniprot
H9IZD2
A0A0L7LFH5
A0A2A4JTJ7
A0A194PX25
A0A194R775
A0A212FIC6
+ More
A0A2A4JSX3 A0A182WBM5 A0A084WME0 A0A182ILK9 A0A182QTC0 A0A182MDK7 A0A1L8E346 A0A1L8E2N7 A0A1J1IV27 A0A182Y2G5 A0A182K9M6 A0A182GHR5 A0A182T5N2 A0A182UA85 A0A182I4M2 A0A182LK98 A0A182X4M4 A0A182PDK3 Q7Q550 A0A182UY22 A0A182FE87 Q16N44 A0A182NI65 A0A336L4B8 A0A182RDJ6 A0A336MRI8 A0A336LN13 W5JKA8 A0A0A1XNW5 A0A1Q3FIK0 A0A0K8V866 W8ANJ8 A0A034VF37 A0A1L8ECM0 A0A0L0BLV1 B4MJH5 A0A1W4VPR4 A0A1I8Q638 A0A1Y1MES8 Q7JWP9 B4MFR7 B4QH39 A0A0J9RI83 B4I828 A0A3B0JP36 B3MGJ5 T1PHD3 A0A1W4X2F3 A0A0R1DYU0 B4P7V9 B4KS89 B4J9Q6 A0A1A9X3D3 A0A2J7PEP5 B0WEF0 Q28XT0 B4GHI8 E2ABP8 A0A067R328 A0A0M4EE59 A0A154P063 A0A2P8XJQ4 A0A088AHC2 D6W899 A0A1B0BPI2 V5GLJ8 A0A2A3E6Q6 A0A1B0G2J5 A0A0L7R3Z0 A0A026X1N5 K7IWN6 A0A158NTA2 A0A195BUR1 E2BXT9 A0A232F9Q1 A0A151K0N0 A0A195E375 E9INY4 A0A3L8D2K2 F4WH12 A0A1A9YC90 A0A195C981 A0A1B6CQQ4 A0A0N0BF34 A0A2S2Q1G6 A0A0C9RF18 U4UKL1
A0A2A4JSX3 A0A182WBM5 A0A084WME0 A0A182ILK9 A0A182QTC0 A0A182MDK7 A0A1L8E346 A0A1L8E2N7 A0A1J1IV27 A0A182Y2G5 A0A182K9M6 A0A182GHR5 A0A182T5N2 A0A182UA85 A0A182I4M2 A0A182LK98 A0A182X4M4 A0A182PDK3 Q7Q550 A0A182UY22 A0A182FE87 Q16N44 A0A182NI65 A0A336L4B8 A0A182RDJ6 A0A336MRI8 A0A336LN13 W5JKA8 A0A0A1XNW5 A0A1Q3FIK0 A0A0K8V866 W8ANJ8 A0A034VF37 A0A1L8ECM0 A0A0L0BLV1 B4MJH5 A0A1W4VPR4 A0A1I8Q638 A0A1Y1MES8 Q7JWP9 B4MFR7 B4QH39 A0A0J9RI83 B4I828 A0A3B0JP36 B3MGJ5 T1PHD3 A0A1W4X2F3 A0A0R1DYU0 B4P7V9 B4KS89 B4J9Q6 A0A1A9X3D3 A0A2J7PEP5 B0WEF0 Q28XT0 B4GHI8 E2ABP8 A0A067R328 A0A0M4EE59 A0A154P063 A0A2P8XJQ4 A0A088AHC2 D6W899 A0A1B0BPI2 V5GLJ8 A0A2A3E6Q6 A0A1B0G2J5 A0A0L7R3Z0 A0A026X1N5 K7IWN6 A0A158NTA2 A0A195BUR1 E2BXT9 A0A232F9Q1 A0A151K0N0 A0A195E375 E9INY4 A0A3L8D2K2 F4WH12 A0A1A9YC90 A0A195C981 A0A1B6CQQ4 A0A0N0BF34 A0A2S2Q1G6 A0A0C9RF18 U4UKL1
Pubmed
19121390
26227816
26354079
22118469
24438588
25244985
+ More
26483478 20966253 12364791 14747013 17210077 17510324 20920257 23761445 25830018 24495485 25348373 26108605 17994087 18057021 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25315136 17550304 15632085 23185243 20798317 24845553 29403074 18362917 19820115 24508170 20075255 21347285 28648823 21282665 30249741 21719571 23537049
26483478 20966253 12364791 14747013 17210077 17510324 20920257 23761445 25830018 24495485 25348373 26108605 17994087 18057021 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25315136 17550304 15632085 23185243 20798317 24845553 29403074 18362917 19820115 24508170 20075255 21347285 28648823 21282665 30249741 21719571 23537049
EMBL
BABH01014279
JTDY01001321
KOB74207.1
NWSH01000701
PCG74712.1
KQ459589
+ More
KPI97563.1 KQ460870 KPJ11721.1 AGBW02008398 OWR53487.1 PCG74714.1 ATLV01024426 KE525352 KFB51384.1 AXCN02000396 AXCM01002092 GFDF01001082 JAV13002.1 GFDF01001081 JAV13003.1 CVRI01000059 CRL03562.1 JXUM01064471 KQ562299 KXJ76207.1 APCN01002331 AAAB01008960 EAA11654.4 CH477840 EAT35763.1 UFQS01001766 UFQT01001766 SSX12269.1 SSX31720.1 SSX12268.1 SSX31719.1 UFQS01000086 UFQT01000086 SSW99090.1 SSX19472.1 ADMH02000963 ETN64561.1 GBXI01006233 GBXI01001712 JAD08059.1 JAD12580.1 GFDL01007648 JAV27397.1 GDHF01017559 GDHF01011804 JAI34755.1 JAI40510.1 GAMC01016181 GAMC01016180 JAB90375.1 GAKP01018784 GAKP01018783 JAC40168.1 GFDG01002496 JAV16303.1 JRES01001695 KNC20903.1 CH963846 EDW72264.2 KRF97520.1 GEZM01037951 JAV81827.1 AE013599 AY113398 AAF46828.1 AAM29403.1 CH940667 EDW57238.1 CM000362 EDX08185.1 CM002911 KMY95758.1 KMY95759.1 CH480824 EDW56753.1 OUUW01000001 SPP75086.1 CH902619 EDV35738.1 KA648089 AFP62718.1 CM000158 KRK00324.1 EDW92114.2 CH933808 EDW08371.1 KRG04080.1 CH916367 EDW01470.1 NEVH01026092 PNF14792.1 DS231908 EDS45577.1 CM000071 EAL26235.2 KRT02932.1 CH479183 EDW35958.1 GL438334 EFN69150.1 KK852945 KDR13470.1 CP012524 ALC41401.1 KQ434786 KZC05251.1 PYGN01001905 PSN32239.1 KQ971307 EFA10916.1 JXJN01017977 JXJN01017978 GALX01003537 JAB64929.1 KZ288365 PBC26849.1 CCAG010013112 KQ414659 KOC65605.1 KK107061 EZA61304.1 ADTU01025763 KQ976409 KYM90948.1 GL451314 EFN79496.1 NNAY01000653 OXU27168.1 KQ981248 KYN44260.1 KQ979701 KYN19598.1 GL764397 EFZ17768.1 QOIP01000014 RLU14735.1 GL888148 EGI66491.1 KQ978072 KYM97270.1 GEDC01021429 JAS15869.1 KQ435819 KOX72498.1 GGMS01002414 MBY71617.1 GBYB01006980 JAG76747.1 KB632399 ERL94614.1
KPI97563.1 KQ460870 KPJ11721.1 AGBW02008398 OWR53487.1 PCG74714.1 ATLV01024426 KE525352 KFB51384.1 AXCN02000396 AXCM01002092 GFDF01001082 JAV13002.1 GFDF01001081 JAV13003.1 CVRI01000059 CRL03562.1 JXUM01064471 KQ562299 KXJ76207.1 APCN01002331 AAAB01008960 EAA11654.4 CH477840 EAT35763.1 UFQS01001766 UFQT01001766 SSX12269.1 SSX31720.1 SSX12268.1 SSX31719.1 UFQS01000086 UFQT01000086 SSW99090.1 SSX19472.1 ADMH02000963 ETN64561.1 GBXI01006233 GBXI01001712 JAD08059.1 JAD12580.1 GFDL01007648 JAV27397.1 GDHF01017559 GDHF01011804 JAI34755.1 JAI40510.1 GAMC01016181 GAMC01016180 JAB90375.1 GAKP01018784 GAKP01018783 JAC40168.1 GFDG01002496 JAV16303.1 JRES01001695 KNC20903.1 CH963846 EDW72264.2 KRF97520.1 GEZM01037951 JAV81827.1 AE013599 AY113398 AAF46828.1 AAM29403.1 CH940667 EDW57238.1 CM000362 EDX08185.1 CM002911 KMY95758.1 KMY95759.1 CH480824 EDW56753.1 OUUW01000001 SPP75086.1 CH902619 EDV35738.1 KA648089 AFP62718.1 CM000158 KRK00324.1 EDW92114.2 CH933808 EDW08371.1 KRG04080.1 CH916367 EDW01470.1 NEVH01026092 PNF14792.1 DS231908 EDS45577.1 CM000071 EAL26235.2 KRT02932.1 CH479183 EDW35958.1 GL438334 EFN69150.1 KK852945 KDR13470.1 CP012524 ALC41401.1 KQ434786 KZC05251.1 PYGN01001905 PSN32239.1 KQ971307 EFA10916.1 JXJN01017977 JXJN01017978 GALX01003537 JAB64929.1 KZ288365 PBC26849.1 CCAG010013112 KQ414659 KOC65605.1 KK107061 EZA61304.1 ADTU01025763 KQ976409 KYM90948.1 GL451314 EFN79496.1 NNAY01000653 OXU27168.1 KQ981248 KYN44260.1 KQ979701 KYN19598.1 GL764397 EFZ17768.1 QOIP01000014 RLU14735.1 GL888148 EGI66491.1 KQ978072 KYM97270.1 GEDC01021429 JAS15869.1 KQ435819 KOX72498.1 GGMS01002414 MBY71617.1 GBYB01006980 JAG76747.1 KB632399 ERL94614.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000075920 UP000030765 UP000075880 UP000075886 UP000075883 UP000183832 UP000076408 UP000075881 UP000069940 UP000249989 UP000075901 UP000075902 UP000075840 UP000075882 UP000076407 UP000075885 UP000007062 UP000075903 UP000069272 UP000008820 UP000075884 UP000075900 UP000000673 UP000037069 UP000007798 UP000192221 UP000095300 UP000000803 UP000008792 UP000000304 UP000001292 UP000268350 UP000007801 UP000095301 UP000192223 UP000002282 UP000009192 UP000001070 UP000091820 UP000235965 UP000002320 UP000001819 UP000008744 UP000000311 UP000027135 UP000092553 UP000076502 UP000245037 UP000005203 UP000007266 UP000092460 UP000242457 UP000092444 UP000053825 UP000053097 UP000002358 UP000005205 UP000078540 UP000008237 UP000215335 UP000078541 UP000078492 UP000279307 UP000007755 UP000092443 UP000078542 UP000053105 UP000030742
UP000075920 UP000030765 UP000075880 UP000075886 UP000075883 UP000183832 UP000076408 UP000075881 UP000069940 UP000249989 UP000075901 UP000075902 UP000075840 UP000075882 UP000076407 UP000075885 UP000007062 UP000075903 UP000069272 UP000008820 UP000075884 UP000075900 UP000000673 UP000037069 UP000007798 UP000192221 UP000095300 UP000000803 UP000008792 UP000000304 UP000001292 UP000268350 UP000007801 UP000095301 UP000192223 UP000002282 UP000009192 UP000001070 UP000091820 UP000235965 UP000002320 UP000001819 UP000008744 UP000000311 UP000027135 UP000092553 UP000076502 UP000245037 UP000005203 UP000007266 UP000092460 UP000242457 UP000092444 UP000053825 UP000053097 UP000002358 UP000005205 UP000078540 UP000008237 UP000215335 UP000078541 UP000078492 UP000279307 UP000007755 UP000092443 UP000078542 UP000053105 UP000030742
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9IZD2
A0A0L7LFH5
A0A2A4JTJ7
A0A194PX25
A0A194R775
A0A212FIC6
+ More
A0A2A4JSX3 A0A182WBM5 A0A084WME0 A0A182ILK9 A0A182QTC0 A0A182MDK7 A0A1L8E346 A0A1L8E2N7 A0A1J1IV27 A0A182Y2G5 A0A182K9M6 A0A182GHR5 A0A182T5N2 A0A182UA85 A0A182I4M2 A0A182LK98 A0A182X4M4 A0A182PDK3 Q7Q550 A0A182UY22 A0A182FE87 Q16N44 A0A182NI65 A0A336L4B8 A0A182RDJ6 A0A336MRI8 A0A336LN13 W5JKA8 A0A0A1XNW5 A0A1Q3FIK0 A0A0K8V866 W8ANJ8 A0A034VF37 A0A1L8ECM0 A0A0L0BLV1 B4MJH5 A0A1W4VPR4 A0A1I8Q638 A0A1Y1MES8 Q7JWP9 B4MFR7 B4QH39 A0A0J9RI83 B4I828 A0A3B0JP36 B3MGJ5 T1PHD3 A0A1W4X2F3 A0A0R1DYU0 B4P7V9 B4KS89 B4J9Q6 A0A1A9X3D3 A0A2J7PEP5 B0WEF0 Q28XT0 B4GHI8 E2ABP8 A0A067R328 A0A0M4EE59 A0A154P063 A0A2P8XJQ4 A0A088AHC2 D6W899 A0A1B0BPI2 V5GLJ8 A0A2A3E6Q6 A0A1B0G2J5 A0A0L7R3Z0 A0A026X1N5 K7IWN6 A0A158NTA2 A0A195BUR1 E2BXT9 A0A232F9Q1 A0A151K0N0 A0A195E375 E9INY4 A0A3L8D2K2 F4WH12 A0A1A9YC90 A0A195C981 A0A1B6CQQ4 A0A0N0BF34 A0A2S2Q1G6 A0A0C9RF18 U4UKL1
A0A2A4JSX3 A0A182WBM5 A0A084WME0 A0A182ILK9 A0A182QTC0 A0A182MDK7 A0A1L8E346 A0A1L8E2N7 A0A1J1IV27 A0A182Y2G5 A0A182K9M6 A0A182GHR5 A0A182T5N2 A0A182UA85 A0A182I4M2 A0A182LK98 A0A182X4M4 A0A182PDK3 Q7Q550 A0A182UY22 A0A182FE87 Q16N44 A0A182NI65 A0A336L4B8 A0A182RDJ6 A0A336MRI8 A0A336LN13 W5JKA8 A0A0A1XNW5 A0A1Q3FIK0 A0A0K8V866 W8ANJ8 A0A034VF37 A0A1L8ECM0 A0A0L0BLV1 B4MJH5 A0A1W4VPR4 A0A1I8Q638 A0A1Y1MES8 Q7JWP9 B4MFR7 B4QH39 A0A0J9RI83 B4I828 A0A3B0JP36 B3MGJ5 T1PHD3 A0A1W4X2F3 A0A0R1DYU0 B4P7V9 B4KS89 B4J9Q6 A0A1A9X3D3 A0A2J7PEP5 B0WEF0 Q28XT0 B4GHI8 E2ABP8 A0A067R328 A0A0M4EE59 A0A154P063 A0A2P8XJQ4 A0A088AHC2 D6W899 A0A1B0BPI2 V5GLJ8 A0A2A3E6Q6 A0A1B0G2J5 A0A0L7R3Z0 A0A026X1N5 K7IWN6 A0A158NTA2 A0A195BUR1 E2BXT9 A0A232F9Q1 A0A151K0N0 A0A195E375 E9INY4 A0A3L8D2K2 F4WH12 A0A1A9YC90 A0A195C981 A0A1B6CQQ4 A0A0N0BF34 A0A2S2Q1G6 A0A0C9RF18 U4UKL1
PDB
4KT1
E-value=1.75985e-19,
Score=239
Ontologies
GO
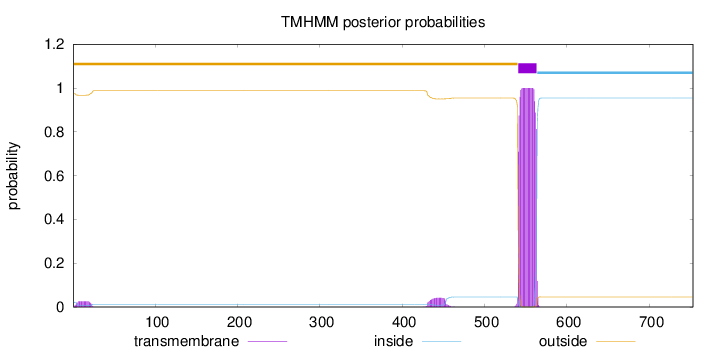
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.992881
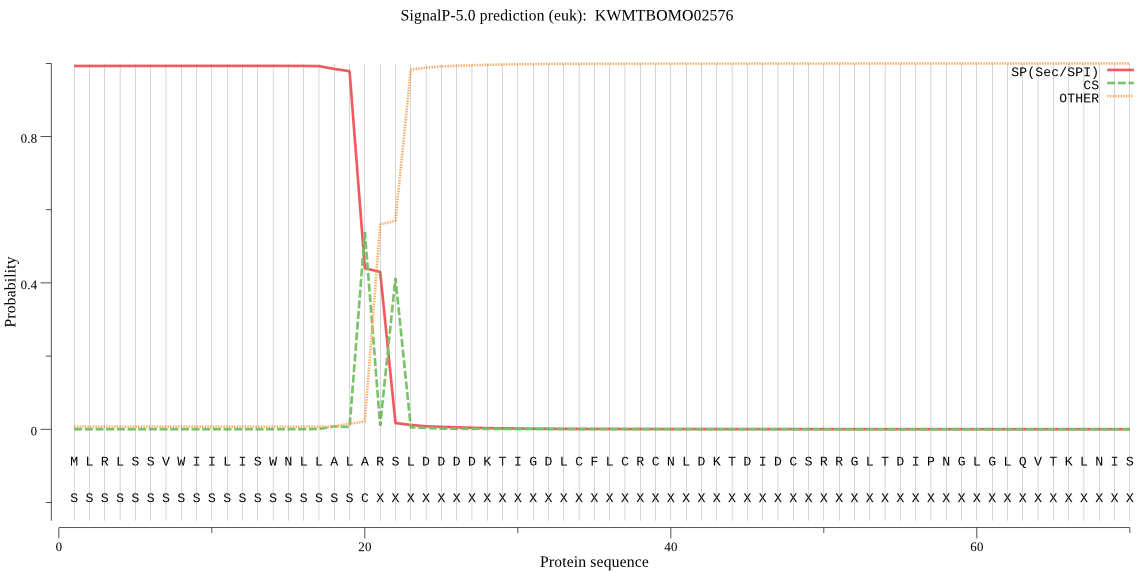
Length:
753
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.77129
Exp number, first 60 AAs:
0.48235
Total prob of N-in:
0.02630
outside
1 - 540
TMhelix
541 - 563
inside
564 - 753
Population Genetic Test Statistics
Pi
28.853052
Theta
20.851589
Tajima's D
0.648995
CLR
0.576428
CSRT
0.56287185640718
Interpretation
Uncertain
