Gene
KWMTBOMO02568
Pre Gene Modal
BGIBMGA002678
Annotation
PREDICTED:_SHC-transforming_protein_1_isoform_X1_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.126 Nuclear Reliability : 1.331
Sequence
CDS
ATGGACTGTGTGTCGTGCCATATTAAAATTAACAATACAAAGTGCGAGGAAAGCGTAAGGCACTACGTGAACCAGACCCCCCCGCCGCGGACCCCGCCCACCGCGCTGCTGCCGAACCACCACACGGACATTTTTGACATGCAGCCATTCACGGCGGCCCCGGTCACGTCGTCGTCGTCCTCTGTGACCTCGGGCACGTCCGCGTCCGCCGAATGCGCGCCGCTCACTGCCGAGGCGCAGGCCGCGCTGCTGGCTAAGGAGCCCTGGTTCCACGGACCTATTTCTAGGACTACTGCTGAAAAGTTGGTAGTTGAGGATGGCGAGTTTTTGGTGCGCGAGTCGACGGCGTGCGCGGGCCAGTTCGTACTGACGGGCGCCAGGAGGGGCCTGCACAAACATCTACTACTCGTCGATCCTATAGGGGTGGTACGAACTAAAGATCGGGTGTTCGACAGTGTACCCCATCTAATCAAGTACCACTGTTCTAACGAGCTGCCCATAGTCTCAGCGGATTCCGCTCTACTACTCCGTAAGCCGGTCCTCAGGTCGTCTTCCTGA
Protein
MDCVSCHIKINNTKCEESVRHYVNQTPPPRTPPTALLPNHHTDIFDMQPFTAAPVTSSSSSVTSGTSASAECAPLTAEAQAALLAKEPWFHGPISRTTAEKLVVEDGEFLVRESTACAGQFVLTGARRGLHKHLLLVDPIGVVRTKDRVFDSVPHLIKYHCSNELPIVSADSALLLRKPVLRSSS
Summary
Uniprot
H9IZJ3
A0A212FK85
A0A2A4J4H4
A0A194PXU0
A0A194R2I2
A0A2J7R7Y0
+ More
A0A1B6K175 A0A067RH42 A0A232FF77 K7J3I3 A0A0C9QLC2 A0A1B6D1S3 A0A182GTT2 A0A1B0EYZ7 E1ZX94 A0A1W4XAS6 A0A1B0CEK1 A0A0M8ZR21 Q16XW4 A0A182WF64 A0A1L8DLM7 A0A1W4XKZ5 A0A182LU83 T1J823 A0A154PM96 A0A0L7QNS6 A0A1Y1MD43 A0A182YPF0 A0A182THQ4 E9IG49 A0A182XC96 A0A182HNJ7 A0A182VBX4 A0A182LPI2 F4WBU3 A0A1Q3F3M8 A0A182QKF9 A0A182MXV9 A0A034WHH7 A0A158NW88 U5EK51 A0A151WN41 A0A084WBD9 A0A195DJ98 B0WGX0 A0A1Q3F3M5 A0A182FPN4 A0A088A1I3 A0A2A3EIH5 A0A182R8P6 A0A182IKU1 A0A0K8V3X7 A0A2M4CRM7 A0A2M3ZAM3 A0A2M4BNW5 A0A026WLQ6 A0A2M4AS25 A0A195FU91 A0A2P2IBB6 F6RER1 A0A1W2WL30 A0A0K8T2V7 A0A0A9VWG0 A0A146LNW6 A0A1D2MSC5 E2B5Y3 A0A2R8QEV1 E7F3T0 E9QCZ8 A0A2G8LMA0 A0A3B4DR52 A0A3B4C1L1 A0A1S3D261 A0A3B4DSH0 A0A336L6Z9 W5LAG0 W8B356
A0A1B6K175 A0A067RH42 A0A232FF77 K7J3I3 A0A0C9QLC2 A0A1B6D1S3 A0A182GTT2 A0A1B0EYZ7 E1ZX94 A0A1W4XAS6 A0A1B0CEK1 A0A0M8ZR21 Q16XW4 A0A182WF64 A0A1L8DLM7 A0A1W4XKZ5 A0A182LU83 T1J823 A0A154PM96 A0A0L7QNS6 A0A1Y1MD43 A0A182YPF0 A0A182THQ4 E9IG49 A0A182XC96 A0A182HNJ7 A0A182VBX4 A0A182LPI2 F4WBU3 A0A1Q3F3M8 A0A182QKF9 A0A182MXV9 A0A034WHH7 A0A158NW88 U5EK51 A0A151WN41 A0A084WBD9 A0A195DJ98 B0WGX0 A0A1Q3F3M5 A0A182FPN4 A0A088A1I3 A0A2A3EIH5 A0A182R8P6 A0A182IKU1 A0A0K8V3X7 A0A2M4CRM7 A0A2M3ZAM3 A0A2M4BNW5 A0A026WLQ6 A0A2M4AS25 A0A195FU91 A0A2P2IBB6 F6RER1 A0A1W2WL30 A0A0K8T2V7 A0A0A9VWG0 A0A146LNW6 A0A1D2MSC5 E2B5Y3 A0A2R8QEV1 E7F3T0 E9QCZ8 A0A2G8LMA0 A0A3B4DR52 A0A3B4C1L1 A0A1S3D261 A0A3B4DSH0 A0A336L6Z9 W5LAG0 W8B356
Pubmed
EMBL
BABH01014311
BABH01014312
AGBW02008106
OWR54151.1
NWSH01003271
PCG66636.1
+ More
KQ459589 KPI97569.1 KQ460870 KPJ11729.1 NEVH01006723 PNF36933.1 GECU01002528 JAT05179.1 KK852685 KDR18487.1 NNAY01000337 OXU29098.1 GBYB01001412 JAG71179.1 GEDC01024757 GEDC01017659 GEDC01014144 GEDC01014123 JAS12541.1 JAS19639.1 JAS23154.1 JAS23175.1 JXUM01087788 KQ563658 KXJ73540.1 AJVK01007412 GL435030 EFN74184.1 AJWK01008943 KQ435971 KOX67779.1 CH477530 EAT39480.1 GFDF01006716 JAV07368.1 AXCM01000254 JH431945 KQ434978 KZC12979.1 KQ414851 KOC60219.1 GEZM01034529 GEZM01034527 JAV83739.1 GL762910 EFZ20455.1 APCN01001937 GL888066 EGI68371.1 GFDL01012885 JAV22160.1 AXCN02002115 GAKP01005181 GAKP01005180 JAC53772.1 ADTU01027850 GANO01001946 JAB57925.1 KQ982911 KYQ49322.1 ATLV01022345 ATLV01022346 KE525331 KFB47533.1 KQ980824 KYN12569.1 DS231930 EDS27264.1 GFDL01012894 JAV22151.1 KZ288241 PBC31274.1 GDHF01018801 JAI33513.1 GGFL01003747 MBW67925.1 GGFM01004828 MBW25579.1 GGFJ01005553 MBW54694.1 KK107154 QOIP01000001 EZA56888.1 RLU27535.1 GGFK01010248 MBW43569.1 KQ981280 KYN43454.1 IACF01005726 LAB71309.1 EAAA01000610 GBRD01006095 JAG59726.1 GBHO01045066 JAF98537.1 GDHC01009028 JAQ09601.1 LJIJ01000643 ODM95625.1 GL445887 EFN88952.1 CR854942 MRZV01000033 PIK61387.1 UFQS01001980 UFQS01002157 UFQT01001980 UFQT01002157 SSX12846.1 SSX13370.1 SSX32288.1 GAMC01011005 JAB95550.1
KQ459589 KPI97569.1 KQ460870 KPJ11729.1 NEVH01006723 PNF36933.1 GECU01002528 JAT05179.1 KK852685 KDR18487.1 NNAY01000337 OXU29098.1 GBYB01001412 JAG71179.1 GEDC01024757 GEDC01017659 GEDC01014144 GEDC01014123 JAS12541.1 JAS19639.1 JAS23154.1 JAS23175.1 JXUM01087788 KQ563658 KXJ73540.1 AJVK01007412 GL435030 EFN74184.1 AJWK01008943 KQ435971 KOX67779.1 CH477530 EAT39480.1 GFDF01006716 JAV07368.1 AXCM01000254 JH431945 KQ434978 KZC12979.1 KQ414851 KOC60219.1 GEZM01034529 GEZM01034527 JAV83739.1 GL762910 EFZ20455.1 APCN01001937 GL888066 EGI68371.1 GFDL01012885 JAV22160.1 AXCN02002115 GAKP01005181 GAKP01005180 JAC53772.1 ADTU01027850 GANO01001946 JAB57925.1 KQ982911 KYQ49322.1 ATLV01022345 ATLV01022346 KE525331 KFB47533.1 KQ980824 KYN12569.1 DS231930 EDS27264.1 GFDL01012894 JAV22151.1 KZ288241 PBC31274.1 GDHF01018801 JAI33513.1 GGFL01003747 MBW67925.1 GGFM01004828 MBW25579.1 GGFJ01005553 MBW54694.1 KK107154 QOIP01000001 EZA56888.1 RLU27535.1 GGFK01010248 MBW43569.1 KQ981280 KYN43454.1 IACF01005726 LAB71309.1 EAAA01000610 GBRD01006095 JAG59726.1 GBHO01045066 JAF98537.1 GDHC01009028 JAQ09601.1 LJIJ01000643 ODM95625.1 GL445887 EFN88952.1 CR854942 MRZV01000033 PIK61387.1 UFQS01001980 UFQS01002157 UFQT01001980 UFQT01002157 SSX12846.1 SSX13370.1 SSX32288.1 GAMC01011005 JAB95550.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000235965
+ More
UP000027135 UP000215335 UP000002358 UP000069940 UP000249989 UP000092462 UP000000311 UP000192223 UP000092461 UP000053105 UP000008820 UP000075920 UP000075883 UP000076502 UP000053825 UP000076408 UP000075902 UP000075840 UP000075903 UP000075882 UP000007755 UP000075886 UP000075884 UP000005205 UP000075809 UP000030765 UP000078492 UP000002320 UP000069272 UP000005203 UP000242457 UP000075900 UP000075880 UP000053097 UP000279307 UP000078541 UP000008144 UP000094527 UP000008237 UP000000437 UP000230750 UP000261440 UP000079169 UP000018467
UP000027135 UP000215335 UP000002358 UP000069940 UP000249989 UP000092462 UP000000311 UP000192223 UP000092461 UP000053105 UP000008820 UP000075920 UP000075883 UP000076502 UP000053825 UP000076408 UP000075902 UP000075840 UP000075903 UP000075882 UP000007755 UP000075886 UP000075884 UP000005205 UP000075809 UP000030765 UP000078492 UP000002320 UP000069272 UP000005203 UP000242457 UP000075900 UP000075880 UP000053097 UP000279307 UP000078541 UP000008144 UP000094527 UP000008237 UP000000437 UP000230750 UP000261440 UP000079169 UP000018467
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IZJ3
A0A212FK85
A0A2A4J4H4
A0A194PXU0
A0A194R2I2
A0A2J7R7Y0
+ More
A0A1B6K175 A0A067RH42 A0A232FF77 K7J3I3 A0A0C9QLC2 A0A1B6D1S3 A0A182GTT2 A0A1B0EYZ7 E1ZX94 A0A1W4XAS6 A0A1B0CEK1 A0A0M8ZR21 Q16XW4 A0A182WF64 A0A1L8DLM7 A0A1W4XKZ5 A0A182LU83 T1J823 A0A154PM96 A0A0L7QNS6 A0A1Y1MD43 A0A182YPF0 A0A182THQ4 E9IG49 A0A182XC96 A0A182HNJ7 A0A182VBX4 A0A182LPI2 F4WBU3 A0A1Q3F3M8 A0A182QKF9 A0A182MXV9 A0A034WHH7 A0A158NW88 U5EK51 A0A151WN41 A0A084WBD9 A0A195DJ98 B0WGX0 A0A1Q3F3M5 A0A182FPN4 A0A088A1I3 A0A2A3EIH5 A0A182R8P6 A0A182IKU1 A0A0K8V3X7 A0A2M4CRM7 A0A2M3ZAM3 A0A2M4BNW5 A0A026WLQ6 A0A2M4AS25 A0A195FU91 A0A2P2IBB6 F6RER1 A0A1W2WL30 A0A0K8T2V7 A0A0A9VWG0 A0A146LNW6 A0A1D2MSC5 E2B5Y3 A0A2R8QEV1 E7F3T0 E9QCZ8 A0A2G8LMA0 A0A3B4DR52 A0A3B4C1L1 A0A1S3D261 A0A3B4DSH0 A0A336L6Z9 W5LAG0 W8B356
A0A1B6K175 A0A067RH42 A0A232FF77 K7J3I3 A0A0C9QLC2 A0A1B6D1S3 A0A182GTT2 A0A1B0EYZ7 E1ZX94 A0A1W4XAS6 A0A1B0CEK1 A0A0M8ZR21 Q16XW4 A0A182WF64 A0A1L8DLM7 A0A1W4XKZ5 A0A182LU83 T1J823 A0A154PM96 A0A0L7QNS6 A0A1Y1MD43 A0A182YPF0 A0A182THQ4 E9IG49 A0A182XC96 A0A182HNJ7 A0A182VBX4 A0A182LPI2 F4WBU3 A0A1Q3F3M8 A0A182QKF9 A0A182MXV9 A0A034WHH7 A0A158NW88 U5EK51 A0A151WN41 A0A084WBD9 A0A195DJ98 B0WGX0 A0A1Q3F3M5 A0A182FPN4 A0A088A1I3 A0A2A3EIH5 A0A182R8P6 A0A182IKU1 A0A0K8V3X7 A0A2M4CRM7 A0A2M3ZAM3 A0A2M4BNW5 A0A026WLQ6 A0A2M4AS25 A0A195FU91 A0A2P2IBB6 F6RER1 A0A1W2WL30 A0A0K8T2V7 A0A0A9VWG0 A0A146LNW6 A0A1D2MSC5 E2B5Y3 A0A2R8QEV1 E7F3T0 E9QCZ8 A0A2G8LMA0 A0A3B4DR52 A0A3B4C1L1 A0A1S3D261 A0A3B4DSH0 A0A336L6Z9 W5LAG0 W8B356
PDB
1MIL
E-value=2.61031e-28,
Score=308
Ontologies
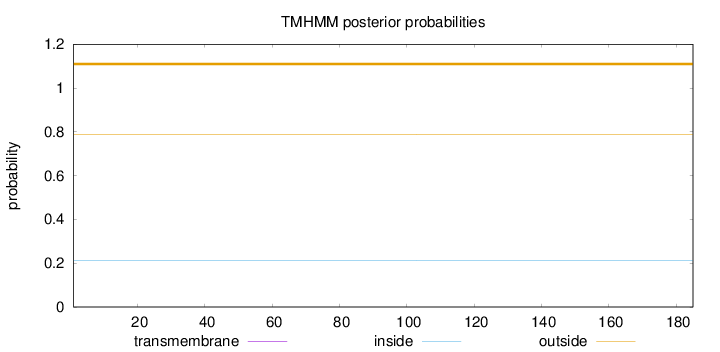
Topology
Length:
185
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000950000000000001
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.21099
outside
1 - 185
Population Genetic Test Statistics
Pi
215.546808
Theta
175.088071
Tajima's D
0.990547
CLR
0.366658
CSRT
0.654617269136543
Interpretation
Uncertain
