Gene
KWMTBOMO02567
Pre Gene Modal
BGIBMGA002611
Annotation
PREDICTED:_lysine-specific_demethylase_6A_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.413 Nuclear Reliability : 1.694
Sequence
CDS
ATGGCTCTGTACGTGTCGCCGGGGTTCTCTCGGGCCGCGGACGCGCACCTTCGGCTGGCGCTCATGTTCAAAGCGCGCCGGCACTGGGCCGCCGCGGCGGTTCACTTCAGGAGGGCTCGCCTCGCGCCGCACCAAGACGCCACCTTCACCCGCCTCGAGCTCAGCTTCCACGCCGCCCACCTGCTGGAGGCGCGGGGCTTGAGGAAGGCCGCCAGGGACGCCTACGAACGCCTCCTCGAGGAACCGAAACTCTCGTCGACACTCAAAGCTGATGTCTGCAGACAATTAGGGTGGTTGTACCATCGGTGCGTGTCTCTGGGCGAGCCCGCAGCGAGGGCACGTGCGGCTATATGGTGTCTCCAACGAGCCGTAGCCGCCGAGCCGGAGTCTGGTGCGGGCCTATACCTTTTAGGGAGGTGTTTCGCTGCTCAAGGCAAAGTCCACGACGCCTTCATCGCGTACAGGAACTCTGTCGAGAAGTCGGAAGGGAACGCCGATACTTGGTGCTCGATCGGTGTCCTATACCAGCAACAGAATCAGCCAATGGACGCGCTCCAAGCGTACATATGTGCCGTGCAGCTTGACAAGGGACATTCGGCGGCGTGGACGAACTTAGGGAGTCTCTATGAGAGCTGTCAGATGCCTCGTGATGCATTCGCGTGCTACACCAATGGAGGTTCGGCCGCGACCGCCAACAACGCGGCGTTGAGACAGCGACTGGCGTTCTTGAAAGCGCACCTGGCACTAGCACCTATGCCCTCAATCACCGGCAAGCGACGTCAGCTGCCTTCCATCGAGGAGGCCTGGAACCTTCCGATCTCCGCGGAGATGTCGTCCCGGGCGCCCAAGTCCGCGCCGCCCCCCTACCCGGGAAAGCGGGACGAGCTCTCGCAACACCAGCTACACACGCTGCACTATCTGCAGAAAAACTCCCACAACCTGACCCCTCAGCAACAAGCGTTGATGCAGCAGCTCTTATCGCAGTACCGTCTCGCTCAGGCAGCAAAAACTCGTGCGGTGACGAAACAGGAAGGTAGCGGCAGCACGACCGGCAGCGACACCACGGAATCGCTTGCCGAAGATTTACTGAAGAAGTTCTCTGACGTACAGCCTGAGATCAAGAAGGAACCGATTAGCTCCGATACAAACACTAGCAACTCGAGCGGCAATGAGGCAGTATTATGCGGCAGACAACCCGTCGTCAAACTCGAGCCGCTCAAGACGGATCCACTTAAACCCGTTACATTCCACATCGGTATGAACTCCAAACAGATTCTCGAGGCTTGCAAAGATAATGCTGGTCCCGCGACCTCTTGGTCGGTACTCGGAGACGGGTGCGGTCCCCCCGAGCCTCCCTCCGTCCCCCCACCGCGTCTCTCCCCCGAACAACTCGCACCCCCCGCTCCTTTCGTGTATGTAGAGTCCAAGCGCGAAGCCTTTTCGCCGCAGCTACAAGATTTCTGCCTGAAACACCCCATCGCCGTGGTGCGCGGACTCACCGCCGCTCTCAAACTCGACCTCGGACTGTTCTCGACGAAGACACTAGTGGAGGCGTGGCCCGACCACGCGGTCGAGGTGCGCACGCAACTCATGCAGTCCGCTGACGAGAACTGGGACGCGACCGGGCGCCGCCGCGTGTGGGCCTGCGCCTCGCACCGCTCCCACACCACCGTGCGCAAGTACGCGCAGTACCAGGCCGGCTCGTTCCAGGAGTCTCTGCGCGAGGAGCGGGAGCGCGCCGCCCCGCTGTCCGACTCGGACTCCGGTCCCGCCAAGCGACGCCGCGCCGCCCGCATGCTCCGCTTCGGCACCAACGTAGACCTCTCAGACGAACGCAAGTGGCGCCCGCAGCTCGCGGAGCTCCAGAAGCTTCCTGCGTTTGCGCGCGTCGCCTCCGCCGCCAACATGCTCTCCCACGTCGGACACGTCATCCTCGGCATGAACACTGTGCAGTTGTACATGAAGGTGCCCGGCAGTCGCACACCAGGACATCAAGAAAATAACAACTTTTGCTCTATCAACCTTAATATCGGCCCCGGCGACTGCGAGTGGTTCGGCGTTCCGGACGCGTACTGGGGAGGCGTGCGCGAGCTGTGCGAGAGACACGGGCTCTCGTACCTGCACGGCTCCTGGTGGCCCGACCCCGACGAGCTGCGCGCGCACGGCGTGCCCGTGTACCGCTTCACGCAGCGGCCCGGCGACCTCGTGTGGGTCAACGCGGGTTGCGTGCACTGGGTGCAGGCCACCGGCTGGTGCAACAACATCGCCTGGAACGTGGGTCCGCTGACGGCGCGCCAGTACACGCTCGCACTCGAGAGATACGAGTGGAACAAAGTGCAGAACTTTAAGTCCATAGTGCCAATGGTACACCTTACGTGGAATTTGGCGCGGAACATCCGCGTCTCGGATCCACGACTGCACCGCGCGATGCGCACGTGTCTGATGCAGACCCTCCGGGCCGCGACGGGCACGCTGAACACGGTGCGCGCGCGCGGGATGCCGATACGGTTCCACGGGCGCGCGCGCGGCGAAGCGTCGCACTACTGCGGCGCGTGCGAGCGCGAGGTGTGGCACGCGCTGCTGGTGCGCGAGCACGAGCGGCGCCACGTGGTGCACTGTCTGCAGTGCGCGCGGCGGGCCAGCCCCTCGCTGCAAGGGTTCCTGTGCCTGGAGGAGCACCACATGGACGAGCTGGCGCAGGTGTACGACGCGTTCACGCTGCACCGGCCGGCGCCGCTGGGTGCGCACGCCGCGCTGCCGCCGGACTGA
Protein
MALYVSPGFSRAADAHLRLALMFKARRHWAAAAVHFRRARLAPHQDATFTRLELSFHAAHLLEARGLRKAARDAYERLLEEPKLSSTLKADVCRQLGWLYHRCVSLGEPAARARAAIWCLQRAVAAEPESGAGLYLLGRCFAAQGKVHDAFIAYRNSVEKSEGNADTWCSIGVLYQQQNQPMDALQAYICAVQLDKGHSAAWTNLGSLYESCQMPRDAFACYTNGGSAATANNAALRQRLAFLKAHLALAPMPSITGKRRQLPSIEEAWNLPISAEMSSRAPKSAPPPYPGKRDELSQHQLHTLHYLQKNSHNLTPQQQALMQQLLSQYRLAQAAKTRAVTKQEGSGSTTGSDTTESLAEDLLKKFSDVQPEIKKEPISSDTNTSNSSGNEAVLCGRQPVVKLEPLKTDPLKPVTFHIGMNSKQILEACKDNAGPATSWSVLGDGCGPPEPPSVPPPRLSPEQLAPPAPFVYVESKREAFSPQLQDFCLKHPIAVVRGLTAALKLDLGLFSTKTLVEAWPDHAVEVRTQLMQSADENWDATGRRRVWACASHRSHTTVRKYAQYQAGSFQESLREERERAAPLSDSDSGPAKRRRAARMLRFGTNVDLSDERKWRPQLAELQKLPAFARVASAANMLSHVGHVILGMNTVQLYMKVPGSRTPGHQENNNFCSINLNIGPGDCEWFGVPDAYWGGVRELCERHGLSYLHGSWWPDPDELRAHGVPVYRFTQRPGDLVWVNAGCVHWVQATGWCNNIAWNVGPLTARQYTLALERYEWNKVQNFKSIVPMVHLTWNLARNIRVSDPRLHRAMRTCLMQTLRAATGTLNTVRARGMPIRFHGRARGEASHYCGACEREVWHALLVREHERRHVVHCLQCARRASPSLQGFLCLEEHHMDELAQVYDAFTLHRPAPLGAHAALPPD
Summary
Uniprot
A0A2H1X0U0
A0A2A4J7S7
A0A2A4J841
A0A212FK90
A0A194R3B6
A0A194Q2E0
+ More
H9IZC6 A0A0J9TKD7 A0A0J9QZY6 M9MRG4 B3N959 B4HWJ4 B4Q8S8 Q9VL07 A0A0J9QZH2 A0A0Q5WNG0 A0A0P9BQ05 A0A0P8ZGK8 A0A0P8XG43 B3MPP9 A0A0P8XGP3 A0A0P8Y4J2 A0A0J9R0T8 M9PCJ7 A0A0J9QZY2 Q76NQ3 A0A0P9A5Y3 A0A2S2QAJ8 A0A0Q5WMI8 A0A0Q5W9U8 A0A0K8TZG9 A0A0Q5WAB1 A0A0Q5WJB9 A0A0K8WE46 A0A0K8W542 A0A034VVC7
H9IZC6 A0A0J9TKD7 A0A0J9QZY6 M9MRG4 B3N959 B4HWJ4 B4Q8S8 Q9VL07 A0A0J9QZH2 A0A0Q5WNG0 A0A0P9BQ05 A0A0P8ZGK8 A0A0P8XG43 B3MPP9 A0A0P8XGP3 A0A0P8Y4J2 A0A0J9R0T8 M9PCJ7 A0A0J9QZY2 Q76NQ3 A0A0P9A5Y3 A0A2S2QAJ8 A0A0Q5WMI8 A0A0Q5W9U8 A0A0K8TZG9 A0A0Q5WAB1 A0A0Q5WJB9 A0A0K8WE46 A0A0K8W542 A0A034VVC7
Pubmed
EMBL
ODYU01012565
SOQ58965.1
NWSH01002753
PCG67574.1
PCG67572.1
AGBW02008106
+ More
OWR54152.1 KQ460870 KPJ11730.1 KQ459589 KPI97570.1 BABH01014313 CM002910 KMY89465.1 KMY89466.1 AE014134 ADV37023.1 CH954177 EDV58494.2 CH480818 EDW52389.1 CM000361 EDX04493.1 KMY89463.1 AY129437 BT010089 AAF52897.3 AAM76179.1 AAQ22558.1 KMY89467.1 KQS70424.1 CH902620 KPU73890.1 KPU73891.1 KPU73887.1 EDV32297.2 KPU73888.1 KPU73889.1 KMY89464.1 AGB92854.1 KMY89468.1 KMY89469.1 KMY89470.1 AAF52898.3 AAN10737.2 KPU73886.1 GGMS01005555 MBY74758.1 KQS70426.1 KQS70423.1 GDHF01032472 GDHF01026708 JAI19842.1 JAI25606.1 KQS70422.1 KQS70421.1 KQS70425.1 GDHF01018734 GDHF01002878 JAI33580.1 JAI49436.1 GDHF01006132 JAI46182.1 GAKP01013167 GAKP01013165 JAC45785.1
OWR54152.1 KQ460870 KPJ11730.1 KQ459589 KPI97570.1 BABH01014313 CM002910 KMY89465.1 KMY89466.1 AE014134 ADV37023.1 CH954177 EDV58494.2 CH480818 EDW52389.1 CM000361 EDX04493.1 KMY89463.1 AY129437 BT010089 AAF52897.3 AAM76179.1 AAQ22558.1 KMY89467.1 KQS70424.1 CH902620 KPU73890.1 KPU73891.1 KPU73887.1 EDV32297.2 KPU73888.1 KPU73889.1 KMY89464.1 AGB92854.1 KMY89468.1 KMY89469.1 KMY89470.1 AAF52898.3 AAN10737.2 KPU73886.1 GGMS01005555 MBY74758.1 KQS70426.1 KQS70423.1 GDHF01032472 GDHF01026708 JAI19842.1 JAI25606.1 KQS70422.1 KQS70421.1 KQS70425.1 GDHF01018734 GDHF01002878 JAI33580.1 JAI49436.1 GDHF01006132 JAI46182.1 GAKP01013167 GAKP01013165 JAC45785.1
Proteomes
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2H1X0U0
A0A2A4J7S7
A0A2A4J841
A0A212FK90
A0A194R3B6
A0A194Q2E0
+ More
H9IZC6 A0A0J9TKD7 A0A0J9QZY6 M9MRG4 B3N959 B4HWJ4 B4Q8S8 Q9VL07 A0A0J9QZH2 A0A0Q5WNG0 A0A0P9BQ05 A0A0P8ZGK8 A0A0P8XG43 B3MPP9 A0A0P8XGP3 A0A0P8Y4J2 A0A0J9R0T8 M9PCJ7 A0A0J9QZY2 Q76NQ3 A0A0P9A5Y3 A0A2S2QAJ8 A0A0Q5WMI8 A0A0Q5W9U8 A0A0K8TZG9 A0A0Q5WAB1 A0A0Q5WJB9 A0A0K8WE46 A0A0K8W542 A0A034VVC7
H9IZC6 A0A0J9TKD7 A0A0J9QZY6 M9MRG4 B3N959 B4HWJ4 B4Q8S8 Q9VL07 A0A0J9QZH2 A0A0Q5WNG0 A0A0P9BQ05 A0A0P8ZGK8 A0A0P8XG43 B3MPP9 A0A0P8XGP3 A0A0P8Y4J2 A0A0J9R0T8 M9PCJ7 A0A0J9QZY2 Q76NQ3 A0A0P9A5Y3 A0A2S2QAJ8 A0A0Q5WMI8 A0A0Q5W9U8 A0A0K8TZG9 A0A0Q5WAB1 A0A0Q5WJB9 A0A0K8WE46 A0A0K8W542 A0A034VVC7
PDB
6G8F
E-value=4.50966e-174,
Score=1573
Ontologies
GO
GO:0046872
GO:0071558
GO:0010468
GO:0008168
GO:0005634
GO:0032452
GO:0044666
GO:0016577
GO:0061086
GO:0051568
GO:0071557
GO:0045498
GO:0005705
GO:0042060
GO:0008285
GO:1902465
GO:0035257
GO:0071480
GO:0010508
GO:0035327
GO:0071390
GO:0035072
GO:1990248
GO:1990841
GO:0045893
GO:0005700
GO:0005515
GO:0016791
GO:0006470
GO:0008138
GO:0004725
GO:0006172
GO:0006631
GO:0006635
GO:0006281
GO:0048384
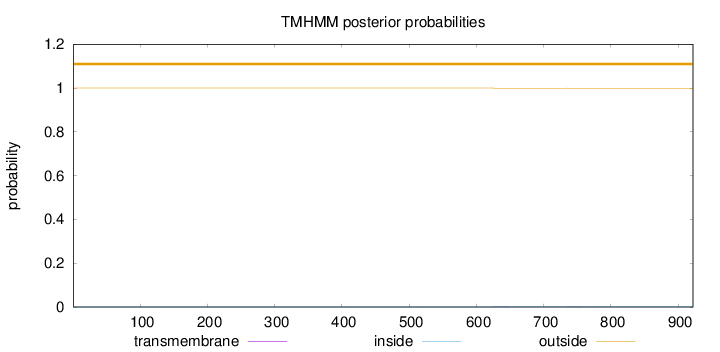
Topology
Length:
922
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01932
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00003
outside
1 - 922
Population Genetic Test Statistics
Pi
177.201231
Theta
194.824996
Tajima's D
-0.755679
CLR
0.941152
CSRT
0.183240837958102
Interpretation
Uncertain
