Gene
KWMTBOMO02565
Pre Gene Modal
BGIBMGA002679
Annotation
PREDICTED:_dual_specificity_phosphatase_DUPD1-like_isoform_X2_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 1.196
Sequence
CDS
ATGAGTTATCGTGACACGAGTTTTGGCTCCTCACCTTCCTCTGTCTTGGGTTCTCTATCATCTCTGCGCTCACGGCCCTTCACTGATGTTTCATACAGCCCAACTCCTGATGTTAATGAAGTGTATCCGGGATTATTTGTTGGAGATGCTGTTGCAGCTAAAGACAAGGCCTTTTTAAGGCGTATGGGCATTAATTATGTCTTGAATACTGCAGAAGGGAAACGCTACACTCAAGTAGACACAGATCATCTCTACTACAGAGACTGTCCTGGTTTGAGATATAAAGGTTTTCAACTTATGGATCTTCCAACTACTGATATTTCAAAGTATTTCCATATTGCTGCCAATTTTATTGATGAGGGAGTATCAAGTGGAGGTCGCGTCCTCGTTCACTGCATGATCGGCGTCTCTCGTTCAGCTACATGTGCACTTGCATTCCTAATGATCAAACGTGGAATGTCCCTCACAGAGGCACTGTGCACTGTACGTGCTCGACGTGACATTCATCCAAATGATGGCTTCCTTCGTCAGCTTCAGCAACTAGAGAGGGACCTACGTATTACACGGGTTCGCTGA
Protein
MSYRDTSFGSSPSSVLGSLSSLRSRPFTDVSYSPTPDVNEVYPGLFVGDAVAAKDKAFLRRMGINYVLNTAEGKRYTQVDTDHLYYRDCPGLRYKGFQLMDLPTTDISKYFHIAANFIDEGVSSGGRVLVHCMIGVSRSATCALAFLMIKRGMSLTEALCTVRARRDIHPNDGFLRQLQQLERDLRITRVR
Summary
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Uniprot
A0A2H1X126
A0A194PX34
A0A194R2I7
A0A2A4JR99
A0A212FK98
H9IZJ4
+ More
A0A0L0BSR8 A0A1J1HH28 B4NCD8 A0A0Q9WTU1 A0A3B0KRR8 Q29GD4 A0A1W4U9Y3 B3NUQ7 A0A3B0KN98 A0A0P9A948 B4R796 B4PWH4 Q9VWN2 A0A3B0KE87 A0A0Q9W869 A0A0R3P615 Q8SXF8 A0A1W4UL84 A0A0Q5T3Z9 B3MZW5 A8JUQ2 A0A0R1EBS7 A0A1B0AZY1 B4MCV2 T1PE17 A0A1I8NF72 A0A1A9XGL3 A0A1A9UZK4 A0A1B0AJ35 A0A0M4FAM3 D3TP49 B4JN12 A0A1I8PNI9 A0A1J1HI08 B4L4D0 A0A0K8VMB2 A0A034WJ16 A0A0K8UVT9 A0A034WI26 A0A336LDT3 A0A336MU89 A0A182T6U7 A0A336LD26 A0A336LBD3 A0A182IV05 U5ERJ3 A0A3F2Z1P0 A0A182RX94 A0A182NFF2 A0A182UWQ1 A0A182TPF9 A0A084W4K3 A0A182XL09 Q7PNB0 A0A182MI49 A0A182PF96 A0A182W6U0 A0A182Y3K6 A0A182L140 B4I6Z3 A0A0R3P1L7 A0A3B0JX08 A0A1W4ULC8 A0A0Q9VZJ8 A0A0R1ECH2 A0A0P8XJ38 A0A3B0KJG6 A0A0R3P196 A0A182QY26 A0A1B0DDB2 A0A1Q3FLR6 A0A1I8NF73 A0A0Q9VZ30 A0A0P8XJ96 A0A1W4U9C1 B0WQL5 X2JEB8 A0A0R1EHP6 A0A1A9X256 T1PM09 A0A1I8NF90 A0A182F159 Q176D4 A0A023EJN1 A0A182GNU6 A0A1J1HLT1 A0A2M4CSS4 A0A2M4A8G3 A0A1Q3FLW3 A0A1S4FDI3 A0A1L8E383
A0A0L0BSR8 A0A1J1HH28 B4NCD8 A0A0Q9WTU1 A0A3B0KRR8 Q29GD4 A0A1W4U9Y3 B3NUQ7 A0A3B0KN98 A0A0P9A948 B4R796 B4PWH4 Q9VWN2 A0A3B0KE87 A0A0Q9W869 A0A0R3P615 Q8SXF8 A0A1W4UL84 A0A0Q5T3Z9 B3MZW5 A8JUQ2 A0A0R1EBS7 A0A1B0AZY1 B4MCV2 T1PE17 A0A1I8NF72 A0A1A9XGL3 A0A1A9UZK4 A0A1B0AJ35 A0A0M4FAM3 D3TP49 B4JN12 A0A1I8PNI9 A0A1J1HI08 B4L4D0 A0A0K8VMB2 A0A034WJ16 A0A0K8UVT9 A0A034WI26 A0A336LDT3 A0A336MU89 A0A182T6U7 A0A336LD26 A0A336LBD3 A0A182IV05 U5ERJ3 A0A3F2Z1P0 A0A182RX94 A0A182NFF2 A0A182UWQ1 A0A182TPF9 A0A084W4K3 A0A182XL09 Q7PNB0 A0A182MI49 A0A182PF96 A0A182W6U0 A0A182Y3K6 A0A182L140 B4I6Z3 A0A0R3P1L7 A0A3B0JX08 A0A1W4ULC8 A0A0Q9VZJ8 A0A0R1ECH2 A0A0P8XJ38 A0A3B0KJG6 A0A0R3P196 A0A182QY26 A0A1B0DDB2 A0A1Q3FLR6 A0A1I8NF73 A0A0Q9VZ30 A0A0P8XJ96 A0A1W4U9C1 B0WQL5 X2JEB8 A0A0R1EHP6 A0A1A9X256 T1PM09 A0A1I8NF90 A0A182F159 Q176D4 A0A023EJN1 A0A182GNU6 A0A1J1HLT1 A0A2M4CSS4 A0A2M4A8G3 A0A1Q3FLW3 A0A1S4FDI3 A0A1L8E383
Pubmed
EMBL
ODYU01012565
SOQ58967.1
KQ459589
KPI97573.1
KQ460870
KPJ11734.1
+ More
NWSH01000735 PCG74511.1 AGBW02008106 OWR54156.1 BABH01014313 JRES01001419 KNC23087.1 CVRI01000004 CRK87187.1 CH964239 EDW82497.1 KRF99626.1 OUUW01000011 SPP86618.1 CH379064 EAL32176.2 CH954180 EDV46654.1 SPP86621.1 CH902635 KPU74837.1 CM000366 EDX18337.1 CM000162 EDX02792.1 AE014298 BT050513 AAF48906.2 ACJ13220.1 SPP86620.1 CH940660 KRF78081.1 KRT07021.1 AY089661 AAL90399.1 KQS30073.1 EDV33916.1 ABW09453.1 KRK06960.1 JXJN01006491 EDW58024.1 KA646949 AFP61578.1 CP012528 ALC49470.1 EZ423201 ADD19477.1 CH916371 EDV92105.1 CRK87186.1 CH933810 EDW07408.1 GDHF01012624 JAI39690.1 GAKP01004620 JAC54332.1 GDHF01021530 JAI30784.1 GAKP01004618 JAC54334.1 UFQS01003878 UFQT01003878 SSX16006.1 SSX35340.1 UFQS01002854 UFQT01002854 SSX14760.1 SSX34154.1 SSX14761.1 SSX34153.1 SSX14759.1 SSX34151.1 GANO01003706 JAB56165.1 ATLV01020343 KE525299 KFB45147.1 AAAB01008964 EAA12467.5 AXCM01003220 CH480823 EDW56091.1 KRT07020.1 SPP86617.1 KRF78082.1 KRK06959.1 KPU74839.1 SPP86619.1 KRT07019.1 AXCN02000817 AJVK01005334 GFDL01006558 JAV28487.1 KRF78083.1 KPU74838.1 DS232041 EDS32890.1 AHN59891.1 KRK06961.1 KA649764 AFP64393.1 CH477390 EAT41967.1 GAPW01004361 JAC09237.1 JXUM01077193 JXUM01077194 JXUM01077195 JXUM01077196 JXUM01077197 JXUM01077198 JXUM01077199 KQ562993 KXJ74690.1 CRK87185.1 GGFL01004195 MBW68373.1 GGFK01003766 MBW37087.1 GFDL01006508 JAV28537.1 GFDF01000985 JAV13099.1
NWSH01000735 PCG74511.1 AGBW02008106 OWR54156.1 BABH01014313 JRES01001419 KNC23087.1 CVRI01000004 CRK87187.1 CH964239 EDW82497.1 KRF99626.1 OUUW01000011 SPP86618.1 CH379064 EAL32176.2 CH954180 EDV46654.1 SPP86621.1 CH902635 KPU74837.1 CM000366 EDX18337.1 CM000162 EDX02792.1 AE014298 BT050513 AAF48906.2 ACJ13220.1 SPP86620.1 CH940660 KRF78081.1 KRT07021.1 AY089661 AAL90399.1 KQS30073.1 EDV33916.1 ABW09453.1 KRK06960.1 JXJN01006491 EDW58024.1 KA646949 AFP61578.1 CP012528 ALC49470.1 EZ423201 ADD19477.1 CH916371 EDV92105.1 CRK87186.1 CH933810 EDW07408.1 GDHF01012624 JAI39690.1 GAKP01004620 JAC54332.1 GDHF01021530 JAI30784.1 GAKP01004618 JAC54334.1 UFQS01003878 UFQT01003878 SSX16006.1 SSX35340.1 UFQS01002854 UFQT01002854 SSX14760.1 SSX34154.1 SSX14761.1 SSX34153.1 SSX14759.1 SSX34151.1 GANO01003706 JAB56165.1 ATLV01020343 KE525299 KFB45147.1 AAAB01008964 EAA12467.5 AXCM01003220 CH480823 EDW56091.1 KRT07020.1 SPP86617.1 KRF78082.1 KRK06959.1 KPU74839.1 SPP86619.1 KRT07019.1 AXCN02000817 AJVK01005334 GFDL01006558 JAV28487.1 KRF78083.1 KPU74838.1 DS232041 EDS32890.1 AHN59891.1 KRK06961.1 KA649764 AFP64393.1 CH477390 EAT41967.1 GAPW01004361 JAC09237.1 JXUM01077193 JXUM01077194 JXUM01077195 JXUM01077196 JXUM01077197 JXUM01077198 JXUM01077199 KQ562993 KXJ74690.1 CRK87185.1 GGFL01004195 MBW68373.1 GGFK01003766 MBW37087.1 GFDL01006508 JAV28537.1 GFDF01000985 JAV13099.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000005204
UP000037069
+ More
UP000183832 UP000007798 UP000268350 UP000001819 UP000192221 UP000008711 UP000007801 UP000000304 UP000002282 UP000000803 UP000008792 UP000092460 UP000095301 UP000092443 UP000078200 UP000092445 UP000092553 UP000001070 UP000095300 UP000009192 UP000075901 UP000075880 UP000076407 UP000075900 UP000075884 UP000075903 UP000075902 UP000030765 UP000007062 UP000075883 UP000075885 UP000075920 UP000076408 UP000075882 UP000001292 UP000075886 UP000092462 UP000002320 UP000091820 UP000069272 UP000008820 UP000069940 UP000249989
UP000183832 UP000007798 UP000268350 UP000001819 UP000192221 UP000008711 UP000007801 UP000000304 UP000002282 UP000000803 UP000008792 UP000092460 UP000095301 UP000092443 UP000078200 UP000092445 UP000092553 UP000001070 UP000095300 UP000009192 UP000075901 UP000075880 UP000076407 UP000075900 UP000075884 UP000075903 UP000075902 UP000030765 UP000007062 UP000075883 UP000075885 UP000075920 UP000076408 UP000075882 UP000001292 UP000075886 UP000092462 UP000002320 UP000091820 UP000069272 UP000008820 UP000069940 UP000249989
Pfam
PF00782 DSPc
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
CDD
ProteinModelPortal
A0A2H1X126
A0A194PX34
A0A194R2I7
A0A2A4JR99
A0A212FK98
H9IZJ4
+ More
A0A0L0BSR8 A0A1J1HH28 B4NCD8 A0A0Q9WTU1 A0A3B0KRR8 Q29GD4 A0A1W4U9Y3 B3NUQ7 A0A3B0KN98 A0A0P9A948 B4R796 B4PWH4 Q9VWN2 A0A3B0KE87 A0A0Q9W869 A0A0R3P615 Q8SXF8 A0A1W4UL84 A0A0Q5T3Z9 B3MZW5 A8JUQ2 A0A0R1EBS7 A0A1B0AZY1 B4MCV2 T1PE17 A0A1I8NF72 A0A1A9XGL3 A0A1A9UZK4 A0A1B0AJ35 A0A0M4FAM3 D3TP49 B4JN12 A0A1I8PNI9 A0A1J1HI08 B4L4D0 A0A0K8VMB2 A0A034WJ16 A0A0K8UVT9 A0A034WI26 A0A336LDT3 A0A336MU89 A0A182T6U7 A0A336LD26 A0A336LBD3 A0A182IV05 U5ERJ3 A0A3F2Z1P0 A0A182RX94 A0A182NFF2 A0A182UWQ1 A0A182TPF9 A0A084W4K3 A0A182XL09 Q7PNB0 A0A182MI49 A0A182PF96 A0A182W6U0 A0A182Y3K6 A0A182L140 B4I6Z3 A0A0R3P1L7 A0A3B0JX08 A0A1W4ULC8 A0A0Q9VZJ8 A0A0R1ECH2 A0A0P8XJ38 A0A3B0KJG6 A0A0R3P196 A0A182QY26 A0A1B0DDB2 A0A1Q3FLR6 A0A1I8NF73 A0A0Q9VZ30 A0A0P8XJ96 A0A1W4U9C1 B0WQL5 X2JEB8 A0A0R1EHP6 A0A1A9X256 T1PM09 A0A1I8NF90 A0A182F159 Q176D4 A0A023EJN1 A0A182GNU6 A0A1J1HLT1 A0A2M4CSS4 A0A2M4A8G3 A0A1Q3FLW3 A0A1S4FDI3 A0A1L8E383
A0A0L0BSR8 A0A1J1HH28 B4NCD8 A0A0Q9WTU1 A0A3B0KRR8 Q29GD4 A0A1W4U9Y3 B3NUQ7 A0A3B0KN98 A0A0P9A948 B4R796 B4PWH4 Q9VWN2 A0A3B0KE87 A0A0Q9W869 A0A0R3P615 Q8SXF8 A0A1W4UL84 A0A0Q5T3Z9 B3MZW5 A8JUQ2 A0A0R1EBS7 A0A1B0AZY1 B4MCV2 T1PE17 A0A1I8NF72 A0A1A9XGL3 A0A1A9UZK4 A0A1B0AJ35 A0A0M4FAM3 D3TP49 B4JN12 A0A1I8PNI9 A0A1J1HI08 B4L4D0 A0A0K8VMB2 A0A034WJ16 A0A0K8UVT9 A0A034WI26 A0A336LDT3 A0A336MU89 A0A182T6U7 A0A336LD26 A0A336LBD3 A0A182IV05 U5ERJ3 A0A3F2Z1P0 A0A182RX94 A0A182NFF2 A0A182UWQ1 A0A182TPF9 A0A084W4K3 A0A182XL09 Q7PNB0 A0A182MI49 A0A182PF96 A0A182W6U0 A0A182Y3K6 A0A182L140 B4I6Z3 A0A0R3P1L7 A0A3B0JX08 A0A1W4ULC8 A0A0Q9VZJ8 A0A0R1ECH2 A0A0P8XJ38 A0A3B0KJG6 A0A0R3P196 A0A182QY26 A0A1B0DDB2 A0A1Q3FLR6 A0A1I8NF73 A0A0Q9VZ30 A0A0P8XJ96 A0A1W4U9C1 B0WQL5 X2JEB8 A0A0R1EHP6 A0A1A9X256 T1PM09 A0A1I8NF90 A0A182F159 Q176D4 A0A023EJN1 A0A182GNU6 A0A1J1HLT1 A0A2M4CSS4 A0A2M4A8G3 A0A1Q3FLW3 A0A1S4FDI3 A0A1L8E383
PDB
2PQ5
E-value=5.14522e-29,
Score=314
Ontologies
GO
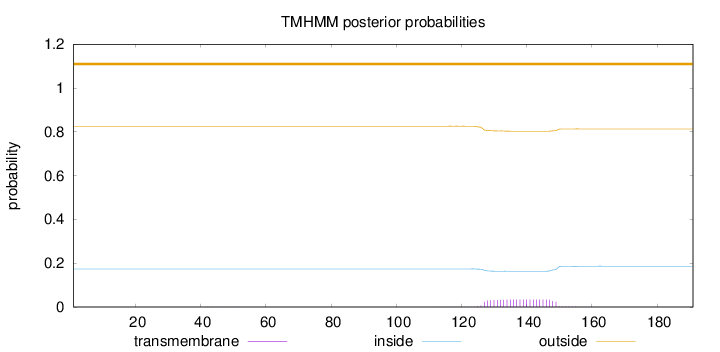
Topology
Length:
191
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.77002
Exp number, first 60 AAs:
0.00331
Total prob of N-in:
0.17399
outside
1 - 191
Population Genetic Test Statistics
Pi
220.321551
Theta
184.657536
Tajima's D
0.616881
CLR
0
CSRT
0.547622618869056
Interpretation
Uncertain
