Pre Gene Modal
BGIBMGA002610
Annotation
39S_ribosomal_protein_L42?_mitochondrial_[Papilio_xuthus]
Full name
Tetraspanin
Location in the cell
Mitochondrial Reliability : 1.734 Nuclear Reliability : 2.11
Sequence
CDS
ATGTCCTATTTAACTCGCAAAATAGCATTAATCCCAAGAGTGGCATTAAGTAGATTCTACAATAAAATAGTGATCACTGACGATGGCTCTACCATTGTAGCTCTTCATCAAGACTATGATTTTCCTTACGAGCATTCAAGAGCTTTACCAGAAGAGACAAGTAAAGATAATACAGTTTTGAAAATGACCGATTTAGATGAAGTTAAAAGAGTCTTCAATGAAATGAAACCTGAATTTGCAAGAAAACAACTATCAAACCTAACATTAACTACTGAACATCGTTGGTACCCTAGAGCTCGAGATAAAAAGGCCAAGAAAACTGAAATGAATAGACCCTATTTGTAA
Protein
MSYLTRKIALIPRVALSRFYNKIVITDDGSTIVALHQDYDFPYEHSRALPEETSKDNTVLKMTDLDEVKRVFNEMKPEFARKQLSNLTLTTEHRWYPRARDKKAKKTEMNRPYL
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9IZC5
A0A194PXU5
A0A194R3C2
A0A2H1X136
A0A2A4JSY9
A0A0L7LUD3
+ More
D6WEX1 A0A067QZS5 A0A1Y1NDI8 V5GFY3 A0A2J7R128 B4GBA3 Q290T1 B4KWL5 A0A2P8Y3T0 T1GGI7 B4MQX6 B4IYJ3 A0A0K8VZV8 A0A034V3Z7 A0A0C9RY45 A0A1I8Q6N7 A0A1B0D1D3 B4LEB4 W8C4Y8 A0A224Y0F3 B4NX43 A0A1W4VD93 A0A0A1XFB0 A0A1B0BI75 A0A1A9YPI2 Q7K0A0 B3N6J3 B4QI84 A0A1A9UJ00 A0A1L8E074 A0A1L8DSA0 A0A1L8EA96 N6TXQ5 A0A023F1T9 B3MD19 A0A1I8MIK6 B4HMC2 A0A1A9WP45 A0A182PTN7 A0A0L0CQM6 A7UTX0 A0A182XMD0 A0A182URA3 A0A1B0G5C8 A0A182KPZ0 A0A182I264 A0A182TDK5 A0A182YEE0 A0A182SL54 A0A1B0A9C4 A0A2M3ZCZ5 A0A182KE98 A0A0T6AY84 A0A0J7K694 A0A2M3ZBH5 A0A182WLB3 A0A182M1Y1 A0A182FHL3 A0A2M4C481 A0A2M4ATL6 A0A2M4ATN8 W5J496 A0A1B6FA13 A0A0N7Z8D7 A0A0V0GB57 E9IWZ8 T1H9Z6 E2A000 A0A2M4C3F3 A0A182NJE4 A0A026WIN3 A0A084WEA1 U5EP49 A0A182R4H8 A0A1B6JN01 A0A195EII3 A0A1J1J7V2 A0A151IJY2 A0A182IWY9 A0A1B6K9T3 A0A195FAW2 E2B311 A0A232ET28 F4WWI5 A0A1B6G6G3 K7IPI5 A0A151WTC1 A0A195B0M5 A0A0P6B1Z2 A0A0P5T4H5 A0A0P5XTS2 J9K879 A0A0P5I7A3
D6WEX1 A0A067QZS5 A0A1Y1NDI8 V5GFY3 A0A2J7R128 B4GBA3 Q290T1 B4KWL5 A0A2P8Y3T0 T1GGI7 B4MQX6 B4IYJ3 A0A0K8VZV8 A0A034V3Z7 A0A0C9RY45 A0A1I8Q6N7 A0A1B0D1D3 B4LEB4 W8C4Y8 A0A224Y0F3 B4NX43 A0A1W4VD93 A0A0A1XFB0 A0A1B0BI75 A0A1A9YPI2 Q7K0A0 B3N6J3 B4QI84 A0A1A9UJ00 A0A1L8E074 A0A1L8DSA0 A0A1L8EA96 N6TXQ5 A0A023F1T9 B3MD19 A0A1I8MIK6 B4HMC2 A0A1A9WP45 A0A182PTN7 A0A0L0CQM6 A7UTX0 A0A182XMD0 A0A182URA3 A0A1B0G5C8 A0A182KPZ0 A0A182I264 A0A182TDK5 A0A182YEE0 A0A182SL54 A0A1B0A9C4 A0A2M3ZCZ5 A0A182KE98 A0A0T6AY84 A0A0J7K694 A0A2M3ZBH5 A0A182WLB3 A0A182M1Y1 A0A182FHL3 A0A2M4C481 A0A2M4ATL6 A0A2M4ATN8 W5J496 A0A1B6FA13 A0A0N7Z8D7 A0A0V0GB57 E9IWZ8 T1H9Z6 E2A000 A0A2M4C3F3 A0A182NJE4 A0A026WIN3 A0A084WEA1 U5EP49 A0A182R4H8 A0A1B6JN01 A0A195EII3 A0A1J1J7V2 A0A151IJY2 A0A182IWY9 A0A1B6K9T3 A0A195FAW2 E2B311 A0A232ET28 F4WWI5 A0A1B6G6G3 K7IPI5 A0A151WTC1 A0A195B0M5 A0A0P6B1Z2 A0A0P5T4H5 A0A0P5XTS2 J9K879 A0A0P5I7A3
Pubmed
19121390
26354079
26227816
18362917
19820115
24845553
+ More
28004739 17994087 15632085 29403074 25348373 24495485 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 23537049 25474469 25315136 26108605 12364791 14747013 17210077 20966253 25244985 20920257 23761445 27129103 21282665 20798317 24508170 30249741 24438588 28648823 21719571 20075255
28004739 17994087 15632085 29403074 25348373 24495485 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 23537049 25474469 25315136 26108605 12364791 14747013 17210077 20966253 25244985 20920257 23761445 27129103 21282665 20798317 24508170 30249741 24438588 28648823 21719571 20075255
EMBL
BABH01014313
KQ459589
KPI97574.1
KQ460870
KPJ11735.1
ODYU01012565
+ More
SOQ58968.1 NWSH01000735 PCG74510.1 JTDY01000106 KOB78831.1 KQ971318 EFA00323.1 KK853060 KDR11909.1 GEZM01005717 JAV95972.1 GALX01005577 JAB62889.1 NEVH01008214 PNF34543.1 CH479181 EDW32205.1 CM000071 EAL25281.3 CH933809 EDW19644.1 PYGN01000957 PSN38916.1 CAQQ02144694 CAQQ02144695 CH963849 EDW74515.1 CH916366 EDV97666.1 GDHF01007945 JAI44369.1 GAKP01022694 JAC36258.1 GBYB01014190 JAG83957.1 AJVK01002574 CH940647 EDW70090.1 GAMC01002099 GAMC01002097 JAC04459.1 GFTR01002006 JAW14420.1 CM000157 EDW89604.1 GBXI01004208 JAD10084.1 JXJN01014858 AE013599 AY070620 AAF58852.1 AAL48091.1 ADV37147.1 CH954177 EDV58162.1 CM000362 CM002911 EDX06444.1 KMY92667.1 GFDF01001993 JAV12091.1 GFDF01004830 JAV09254.1 GFDG01003306 JAV15493.1 APGK01050872 KB741178 KB632256 ENN73161.1 ERL90673.1 GBBI01003424 JAC15288.1 CH902619 EDV36334.1 CH480816 EDW47199.1 JRES01000160 KNC33739.1 AAAB01008960 EDO63744.1 CCAG010003857 APCN01000123 GGFM01005614 MBW26365.1 LJIG01022533 KRT80076.1 LBMM01012762 KMQ86008.1 GGFM01005069 MBW25820.1 AXCM01002346 GGFJ01010667 MBW59808.1 GGFK01010815 MBW44136.1 GGFK01010816 MBW44137.1 ADMH02002096 ETN59252.1 GECZ01022683 JAS47086.1 GDKW01003635 JAI52960.1 GECL01001040 JAP05084.1 GL766616 EFZ14878.1 ACPB03024744 GL435531 EFN73240.1 GGFJ01010691 MBW59832.1 KK107183 QOIP01000011 EZA55880.1 RLU16789.1 ATLV01023198 KE525341 KFB48545.1 GANO01003825 JAB56046.1 GECU01007151 JAT00556.1 KQ978881 KYN27707.1 CVRI01000075 CRL08471.1 KQ977294 KYN03957.1 GEBQ01031768 JAT08209.1 KQ981693 KYN37760.1 GL445250 EFN89980.1 NNAY01002357 OXU21477.1 GL888406 EGI61438.1 GECZ01011741 JAS58028.1 KQ982762 KYQ51078.1 KQ976692 KYM77847.1 GDIP01021580 JAM82135.1 GDIP01131160 JAL72554.1 GDIP01068761 LRGB01000763 JAM34954.1 KZS16112.1 ABLF02036611 GDIQ01223914 JAK27811.1
SOQ58968.1 NWSH01000735 PCG74510.1 JTDY01000106 KOB78831.1 KQ971318 EFA00323.1 KK853060 KDR11909.1 GEZM01005717 JAV95972.1 GALX01005577 JAB62889.1 NEVH01008214 PNF34543.1 CH479181 EDW32205.1 CM000071 EAL25281.3 CH933809 EDW19644.1 PYGN01000957 PSN38916.1 CAQQ02144694 CAQQ02144695 CH963849 EDW74515.1 CH916366 EDV97666.1 GDHF01007945 JAI44369.1 GAKP01022694 JAC36258.1 GBYB01014190 JAG83957.1 AJVK01002574 CH940647 EDW70090.1 GAMC01002099 GAMC01002097 JAC04459.1 GFTR01002006 JAW14420.1 CM000157 EDW89604.1 GBXI01004208 JAD10084.1 JXJN01014858 AE013599 AY070620 AAF58852.1 AAL48091.1 ADV37147.1 CH954177 EDV58162.1 CM000362 CM002911 EDX06444.1 KMY92667.1 GFDF01001993 JAV12091.1 GFDF01004830 JAV09254.1 GFDG01003306 JAV15493.1 APGK01050872 KB741178 KB632256 ENN73161.1 ERL90673.1 GBBI01003424 JAC15288.1 CH902619 EDV36334.1 CH480816 EDW47199.1 JRES01000160 KNC33739.1 AAAB01008960 EDO63744.1 CCAG010003857 APCN01000123 GGFM01005614 MBW26365.1 LJIG01022533 KRT80076.1 LBMM01012762 KMQ86008.1 GGFM01005069 MBW25820.1 AXCM01002346 GGFJ01010667 MBW59808.1 GGFK01010815 MBW44136.1 GGFK01010816 MBW44137.1 ADMH02002096 ETN59252.1 GECZ01022683 JAS47086.1 GDKW01003635 JAI52960.1 GECL01001040 JAP05084.1 GL766616 EFZ14878.1 ACPB03024744 GL435531 EFN73240.1 GGFJ01010691 MBW59832.1 KK107183 QOIP01000011 EZA55880.1 RLU16789.1 ATLV01023198 KE525341 KFB48545.1 GANO01003825 JAB56046.1 GECU01007151 JAT00556.1 KQ978881 KYN27707.1 CVRI01000075 CRL08471.1 KQ977294 KYN03957.1 GEBQ01031768 JAT08209.1 KQ981693 KYN37760.1 GL445250 EFN89980.1 NNAY01002357 OXU21477.1 GL888406 EGI61438.1 GECZ01011741 JAS58028.1 KQ982762 KYQ51078.1 KQ976692 KYM77847.1 GDIP01021580 JAM82135.1 GDIP01131160 JAL72554.1 GDIP01068761 LRGB01000763 JAM34954.1 KZS16112.1 ABLF02036611 GDIQ01223914 JAK27811.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000037510
UP000007266
+ More
UP000027135 UP000235965 UP000008744 UP000001819 UP000009192 UP000245037 UP000015102 UP000007798 UP000001070 UP000095300 UP000092462 UP000008792 UP000002282 UP000192221 UP000092460 UP000092443 UP000000803 UP000008711 UP000000304 UP000078200 UP000019118 UP000030742 UP000007801 UP000095301 UP000001292 UP000091820 UP000075885 UP000037069 UP000007062 UP000076407 UP000075903 UP000092444 UP000075882 UP000075840 UP000075902 UP000076408 UP000075901 UP000092445 UP000075881 UP000036403 UP000075920 UP000075883 UP000069272 UP000000673 UP000015103 UP000000311 UP000075884 UP000053097 UP000279307 UP000030765 UP000075900 UP000078492 UP000183832 UP000078542 UP000075880 UP000078541 UP000008237 UP000215335 UP000007755 UP000002358 UP000075809 UP000078540 UP000076858 UP000007819
UP000027135 UP000235965 UP000008744 UP000001819 UP000009192 UP000245037 UP000015102 UP000007798 UP000001070 UP000095300 UP000092462 UP000008792 UP000002282 UP000192221 UP000092460 UP000092443 UP000000803 UP000008711 UP000000304 UP000078200 UP000019118 UP000030742 UP000007801 UP000095301 UP000001292 UP000091820 UP000075885 UP000037069 UP000007062 UP000076407 UP000075903 UP000092444 UP000075882 UP000075840 UP000075902 UP000076408 UP000075901 UP000092445 UP000075881 UP000036403 UP000075920 UP000075883 UP000069272 UP000000673 UP000015103 UP000000311 UP000075884 UP000053097 UP000279307 UP000030765 UP000075900 UP000078492 UP000183832 UP000078542 UP000075880 UP000078541 UP000008237 UP000215335 UP000007755 UP000002358 UP000075809 UP000078540 UP000076858 UP000007819
PRIDE
Interpro
SUPFAM
SSF48652
SSF48652
Gene 3D
ProteinModelPortal
H9IZC5
A0A194PXU5
A0A194R3C2
A0A2H1X136
A0A2A4JSY9
A0A0L7LUD3
+ More
D6WEX1 A0A067QZS5 A0A1Y1NDI8 V5GFY3 A0A2J7R128 B4GBA3 Q290T1 B4KWL5 A0A2P8Y3T0 T1GGI7 B4MQX6 B4IYJ3 A0A0K8VZV8 A0A034V3Z7 A0A0C9RY45 A0A1I8Q6N7 A0A1B0D1D3 B4LEB4 W8C4Y8 A0A224Y0F3 B4NX43 A0A1W4VD93 A0A0A1XFB0 A0A1B0BI75 A0A1A9YPI2 Q7K0A0 B3N6J3 B4QI84 A0A1A9UJ00 A0A1L8E074 A0A1L8DSA0 A0A1L8EA96 N6TXQ5 A0A023F1T9 B3MD19 A0A1I8MIK6 B4HMC2 A0A1A9WP45 A0A182PTN7 A0A0L0CQM6 A7UTX0 A0A182XMD0 A0A182URA3 A0A1B0G5C8 A0A182KPZ0 A0A182I264 A0A182TDK5 A0A182YEE0 A0A182SL54 A0A1B0A9C4 A0A2M3ZCZ5 A0A182KE98 A0A0T6AY84 A0A0J7K694 A0A2M3ZBH5 A0A182WLB3 A0A182M1Y1 A0A182FHL3 A0A2M4C481 A0A2M4ATL6 A0A2M4ATN8 W5J496 A0A1B6FA13 A0A0N7Z8D7 A0A0V0GB57 E9IWZ8 T1H9Z6 E2A000 A0A2M4C3F3 A0A182NJE4 A0A026WIN3 A0A084WEA1 U5EP49 A0A182R4H8 A0A1B6JN01 A0A195EII3 A0A1J1J7V2 A0A151IJY2 A0A182IWY9 A0A1B6K9T3 A0A195FAW2 E2B311 A0A232ET28 F4WWI5 A0A1B6G6G3 K7IPI5 A0A151WTC1 A0A195B0M5 A0A0P6B1Z2 A0A0P5T4H5 A0A0P5XTS2 J9K879 A0A0P5I7A3
D6WEX1 A0A067QZS5 A0A1Y1NDI8 V5GFY3 A0A2J7R128 B4GBA3 Q290T1 B4KWL5 A0A2P8Y3T0 T1GGI7 B4MQX6 B4IYJ3 A0A0K8VZV8 A0A034V3Z7 A0A0C9RY45 A0A1I8Q6N7 A0A1B0D1D3 B4LEB4 W8C4Y8 A0A224Y0F3 B4NX43 A0A1W4VD93 A0A0A1XFB0 A0A1B0BI75 A0A1A9YPI2 Q7K0A0 B3N6J3 B4QI84 A0A1A9UJ00 A0A1L8E074 A0A1L8DSA0 A0A1L8EA96 N6TXQ5 A0A023F1T9 B3MD19 A0A1I8MIK6 B4HMC2 A0A1A9WP45 A0A182PTN7 A0A0L0CQM6 A7UTX0 A0A182XMD0 A0A182URA3 A0A1B0G5C8 A0A182KPZ0 A0A182I264 A0A182TDK5 A0A182YEE0 A0A182SL54 A0A1B0A9C4 A0A2M3ZCZ5 A0A182KE98 A0A0T6AY84 A0A0J7K694 A0A2M3ZBH5 A0A182WLB3 A0A182M1Y1 A0A182FHL3 A0A2M4C481 A0A2M4ATL6 A0A2M4ATN8 W5J496 A0A1B6FA13 A0A0N7Z8D7 A0A0V0GB57 E9IWZ8 T1H9Z6 E2A000 A0A2M4C3F3 A0A182NJE4 A0A026WIN3 A0A084WEA1 U5EP49 A0A182R4H8 A0A1B6JN01 A0A195EII3 A0A1J1J7V2 A0A151IJY2 A0A182IWY9 A0A1B6K9T3 A0A195FAW2 E2B311 A0A232ET28 F4WWI5 A0A1B6G6G3 K7IPI5 A0A151WTC1 A0A195B0M5 A0A0P6B1Z2 A0A0P5T4H5 A0A0P5XTS2 J9K879 A0A0P5I7A3
PDB
6NU3
E-value=4.36031e-07,
Score=122
Ontologies
PANTHER
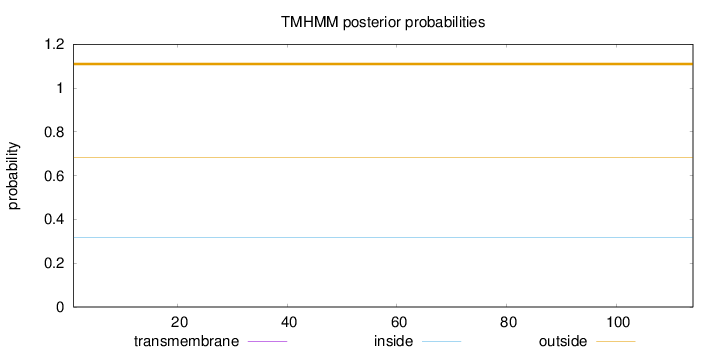
Topology
Subcellular location
Membrane
Length:
114
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00145
Exp number, first 60 AAs:
0.00145
Total prob of N-in:
0.31796
outside
1 - 114
Population Genetic Test Statistics
Pi
19.571966
Theta
29.316824
Tajima's D
-0.786144
CLR
0
CSRT
0.179891005449728
Interpretation
Uncertain
