Gene
KWMTBOMO02560 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002682
Annotation
PREDICTED:_uncharacterized_protein_LOC101739478_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.675 Nuclear Reliability : 1.162
Sequence
CDS
ATGCAAAGAATAGATATGGATATTATCGATGTGTTGCTTCAAGCTGGGTTGATTACTTCAGCCTTGGTTTTGTATTTGTTATTCATACGTAAACAACCGGCACGTAGAGGAGAATATAAATTGCCAGGCTGGAACTACCCCTTGAAGCTTTTGCTAGCTAGATATGCTGTGAAGCGATGGAAAGCTGGTTTACACTCAATAGTCGGAGATGAGTCACCACGTGGTCCACAACACGAAGGCTGGGACTCCATTTCCATTCGCACTGCAGCGCCTAATGGAGTCACGGTGCTTTTGGGAGTTCGGAAATTATGTGGGCGCCAACCCATGGCTGAAATAATTGTGTATATAAAATTAAAAGACGGCACATCTTACGAACTGATGGATTGTCCCGACACACTGGTTGGAGCATGGCTATCAACAGCAGATGGATGGCGTGCTGGCGGACTGAAGATACAAATTTTAGAGCCCCGTTGTCGAATGAGAATTTTATACAACGGTCTTTTAAAAAGAACAACAGACAATGTTACTCAACATGTAACGTTTAATTTAACGTGGACCTCGAGTTCTGTTCCCGTAGAGCATCCCAAAGACTGGAACGAGAAATTAGCGGCACAAACTTTGGCTTTGGAACCATGGCGAGATAGTCAATGGTCGTCGATGCTAGGTAAATGGGAAGAAGGAAGTTGGCTTCAGTGGGGAACCGTGCAAGGACGATTCCAGTCGTTTACTGCGGAAGGCGCTCTTGACACGACTGAATACCTGCAAACTCGAGGAGTGAAAGAGAGGCATTGGGCGCCGCACGCATATGAGGGTACACGGAGATCTTTTTCTATAACGGCATCAGCAAGAGATGGTACCGCTGTCGTGCTACGAGGAGTGTCATACAAAAAGGTTTTGACTCAGTGCTTGTCAGGTTGTGTCAGACTCCCTGATTACAGAGTACTGACTATTACGGATACTGATTTGATCATGTCCGATTTTTGCGAAACTCCTGGTGGAATACCAAGTGCTTACACCATAAACATACACACGAAGGAACGTTCTCTCAAAGTTGTACTCCGCATCGGTGAGGAGGGATGTACGGCTTATAGTGGAGTTCCGTACCAACAGGAACTGGTGTACCGCGCTATTGTCGCTGATATCGACGGGGAAGCGGGCTCTGGTATCTTGGAGCTTGGATATCTGCCTAAAGCCATTGCTCAGCCATCAGTCCGTCTCACTCCAGCGCCCATTTTGAAATGGTTGTTAAAAAAAGATGCTGGTTTCGTTGGCTATTGTTTGTCGTTCGAAGAACGTGCTGCAACGTGTACTGATTACGTCGGCGGGAAAGGAGCATCCTTGGCATTACTAGCCTCCGTACAGAATGATGAGGGATACCGGGTGCCACCGGGTTTTTGTATAACAACTAAAGCACTGGAGAAACATCTTCAATTGCATACTGAACTGAAAGCAGCTATACAAGATATAGAGGCAGCTAATGAAAATTATGAGGAGGCTTACTTCAAAGAACGATGTCAGAAAGTTTCATCATTATTCTTGGCTTTAGAATTAAGCGCAGATATAAAGAAAGACATATTAGAATATATGCAGGAGTTGAGGAGCAAATCCGCACAGTTGAAAGACGCACAGGAGTTGCGATTTGCCGTTCGATCTTCAGCGGTCGGAGAAGACAGCGAGGCGTTATCTGCTGCTGGACAGAACGAAACAATTTTGGGTTGTGTCACCGACGATGACGTGATACGAGCGGTCCAGAAGTGCTGGGGCTCCATGTTTGAGTTCACAAGTACCTATTATCGCAGACAGAACGGCCAAGTATGTTTATGCGGAGGCGGGGTCGTGGTGCAGACGCTGGTGTCGCCTCGCGTCGCCGGCGTCATGTTCACTCGACACCCTGACGCTGGAGATCCTTCGAGGCTACTCATCACCGCAAATTATGGATTGGGAGAAAGCGTCGTCTCCGGTACCGTAGAGCCCGACACGATAATCGTGAAGCGAGAACCAAACGGCGTCCTCACTATCCAGAAAAGGGAACTAGGATCTAAAACGCGACGCCACGTCGCGTCATCTAGCGGCGGAGTCATTACAGAAGACGTTCCGGAAAAAGAAAGAAGTGTTGCATGTTTGAATGACACCGAAGTACTTAAGCTAGCTCGACTAGGTGTTGTACAAGAGGAACTTTGGGGGGCCGGGAGGGATATAGAGTGGGCTATAAGTGGCGATCAAGTGTATTTATTGCAAGCGCGGCCGATTACGTCTTTAGAACGATGGACTGAAGAAGAGCTACTACACGAAGTCGACTCTCCGATTATGGCAGATAACGAACTGACCACATTCGGCAACACTGGAGAGGTTCTTCCGAAGCCTGTAACTCCGCTAACCTACGATTTGGTGATACGTCCGCTGATATGGAGCATGGACCGAGCCATAATCACAAACGGCAGCCAGTACGACCACAGTCTAACGCTCAGCCACTACAGATGCGCTATATCTTTGTATAATTCGGTTTATCGAAGAGTACCAAAAAAGCTAGACACTAACATACGAATGATGGAAATGGCCATAAACGGACACAAAATAGCGGACGAAAACATTCACAAGACCGCGCTGATTCGCAGAAAACCGCACTGGACTGATCGAATAAGGCTTATTCATCATATGATTATGTCTATATTGACTTCGAAATGGAAACTAAACGATACAGTTAAGAAAGCGAAAGATCTAGAAGTTGGAACGAACGCCAAAGAACCAATCGAATTACTGGAAAGTATTGCTGAATGTGAAGATTTGATTGGCGAGCTGACGTACAACCACAGCGTGACCACCGTGGCCAGCACGTCCAGTCAGATTATAGCTATGACCATACTCATGGAAGGGAGTGCAGACTTCACTCCAGAACAATGTAATGAGATCAGCAAATTGCTAAGCTCCGGAGATGTGTTATCAGCAGAGGTTCCACAGTGTTTGGCAAACTTAACTAGGCAATTAGAAAAATCCCGGCTGATGGACGAATTTCGTGCCCAAGACCCGAAGTACGCATTGAGATGGTTGAAAAGGAATCTCCCTCATATTTACCTAGACATTTGCACATTCCTCGAGCACCACGGATACAGGGCTATCATGGAGTTTGATTTATACACGAAGCCATGGATATTGGTCCCGGAAGAAATGATGAAAATACTACAAAACCTACGTTCTTCGCCGGAGGAACCACAACAATCTAAGACAACAGACGAAGTCATCGCTTCCTTGACCACCCCTAAGAAGAACAGCACCAGGAAAGTCCTACGATGGGTGTTGCCTCTCTGCAGGAGAGCGGTCCGCCACCGCGAGGGCACGAAAGCGCACCTCATTCTGGCTGCACATAAAATGCGATTGGCGTTATTACGGCTCGGCGACATGTTGGTACGCCAATGGTATCTGCCGCATCCAGACTTGGTATTTTTCTTCAGGCTGGGCGAACTCCAAGAGTACGTTAGGAAAAGAGATCCGGTGTTGTTAAAGAAGGCAATCCAGCGCCAGCAGTATTACCCGAAATGGTGCAAGTTGAAATTCAACGAACTGAATAATGGATGGATAGCGCCTCTAGAAGCAAAAGGTCCTTCGGTCAGGGCCGGTGACGTCAGAATCGAAGCGACCTCCGTCTGCGGCGGAGAGGTAGTGGCCAGGGCCTGTGTCGTCAAAGAACTTTCTGAAGTAAGAATCATCTGCTAA
Protein
MQRIDMDIIDVLLQAGLITSALVLYLLFIRKQPARRGEYKLPGWNYPLKLLLARYAVKRWKAGLHSIVGDESPRGPQHEGWDSISIRTAAPNGVTVLLGVRKLCGRQPMAEIIVYIKLKDGTSYELMDCPDTLVGAWLSTADGWRAGGLKIQILEPRCRMRILYNGLLKRTTDNVTQHVTFNLTWTSSSVPVEHPKDWNEKLAAQTLALEPWRDSQWSSMLGKWEEGSWLQWGTVQGRFQSFTAEGALDTTEYLQTRGVKERHWAPHAYEGTRRSFSITASARDGTAVVLRGVSYKKVLTQCLSGCVRLPDYRVLTITDTDLIMSDFCETPGGIPSAYTINIHTKERSLKVVLRIGEEGCTAYSGVPYQQELVYRAIVADIDGEAGSGILELGYLPKAIAQPSVRLTPAPILKWLLKKDAGFVGYCLSFEERAATCTDYVGGKGASLALLASVQNDEGYRVPPGFCITTKALEKHLQLHTELKAAIQDIEAANENYEEAYFKERCQKVSSLFLALELSADIKKDILEYMQELRSKSAQLKDAQELRFAVRSSAVGEDSEALSAAGQNETILGCVTDDDVIRAVQKCWGSMFEFTSTYYRRQNGQVCLCGGGVVVQTLVSPRVAGVMFTRHPDAGDPSRLLITANYGLGESVVSGTVEPDTIIVKREPNGVLTIQKRELGSKTRRHVASSSGGVITEDVPEKERSVACLNDTEVLKLARLGVVQEELWGAGRDIEWAISGDQVYLLQARPITSLERWTEEELLHEVDSPIMADNELTTFGNTGEVLPKPVTPLTYDLVIRPLIWSMDRAIITNGSQYDHSLTLSHYRCAISLYNSVYRRVPKKLDTNIRMMEMAINGHKIADENIHKTALIRRKPHWTDRIRLIHHMIMSILTSKWKLNDTVKKAKDLEVGTNAKEPIELLESIAECEDLIGELTYNHSVTTVASTSSQIIAMTILMEGSADFTPEQCNEISKLLSSGDVLSAEVPQCLANLTRQLEKSRLMDEFRAQDPKYALRWLKRNLPHIYLDICTFLEHHGYRAIMEFDLYTKPWILVPEEMMKILQNLRSSPEEPQQSKTTDEVIASLTTPKKNSTRKVLRWVLPLCRRAVRHREGTKAHLILAAHKMRLALLRLGDMLVRQWYLPHPDLVFFFRLGELQEYVRKRDPVLLKKAIQRQQYYPKWCKLKFNELNNGWIAPLEAKGPSVRAGDVRIEATSVCGGEVVARACVVKELSEVRIIC
Summary
Uniprot
A0A2A4JRX2
A0A2H1WY32
A0A194QV09
A0A194PXV0
A0A3L8DLP2
A0A026X3J0
+ More
A0A151ILI8 A0A158NVZ5 A0A151WSS2 A0A2J7Q877 A0A2J7Q868 A0A151HY46 A0A195ES97 K7JPF5 A0A0N0BF09 E2BY46 A0A310S601 A0A139WGR7 A0A0L7QV50 A0A0P5WPZ2 A0A2J7Q876 A0A0P5H5T3 A0A0P5WRL6 A0A0N8D7Q2 A0A0P5AWX9 A0A0P5RGS6 A0A0P5ATU0 A0A0P5A875 A0A0P6DU69 A0A0P5IFV5 A0A0P6DYB3 A0A0P5JS45 A0A154P524 A0A0P5IDP6 A0A0P5PYV7 A0A0P6GB77 A0A0P5KRB8 E9GD24 A0A0P5S2P4 A0A0P5R3H5 A0A0P5GYA6 A0A0N8BPI2 A0A0P5PLP8 A0A0P5JZ33 A0A0P6HEP5 A0A067QI31 A0A0P5K061 A0A0P5LPF8 A0A0P5SEQ5 A0A0P5LD82 A0A0P5VUB4 A0A0P5JBM5 A0A0P5L5V1 A0A0P6DB64 A0A0P5KKB0 A0A0N7ZUZ1 A0A0P5PC38 A0A0P5SY78 A0A0P6CYU6 A0A0P5N754 A0A0P6E3W7 A0A0P5KYN3 A0A0N8BZ42 A0A0P5KRY9 A0A0P6DHG3 A0A0P5L176 A0A0P5LJY0 A0A0P5MXZ2 A0A0P5LIH6 A0A0P5JVC6 A0A0P5P1H8 A0A0P5NG43 A0A0N8DVZ9 A0A0P6DW90 A0A0P5QRW2 A0A0P5VZK7 A0A0P5W213 A0A0P4Z5F4 A0A0P5Q110 A0A0P5UHB8 A0A0P4ZW92 A0A0N8AVA9 A0A2J7Q872 A0A0P5ID55 A0A0P5ZET5 A0A0P6ERF7 A0A0P5JJ67 A0A0P5BNP1 A0A0P5X2K1 A0A0P5BR09 A0A0P5VZH1 A0A0P4ZVU9 A0A2J7Q871 A0A0P5Q230 A0A0P5WVY8 A0A0N8AES1 A0A0P5PMB6 A0A0P5M0M0
A0A151ILI8 A0A158NVZ5 A0A151WSS2 A0A2J7Q877 A0A2J7Q868 A0A151HY46 A0A195ES97 K7JPF5 A0A0N0BF09 E2BY46 A0A310S601 A0A139WGR7 A0A0L7QV50 A0A0P5WPZ2 A0A2J7Q876 A0A0P5H5T3 A0A0P5WRL6 A0A0N8D7Q2 A0A0P5AWX9 A0A0P5RGS6 A0A0P5ATU0 A0A0P5A875 A0A0P6DU69 A0A0P5IFV5 A0A0P6DYB3 A0A0P5JS45 A0A154P524 A0A0P5IDP6 A0A0P5PYV7 A0A0P6GB77 A0A0P5KRB8 E9GD24 A0A0P5S2P4 A0A0P5R3H5 A0A0P5GYA6 A0A0N8BPI2 A0A0P5PLP8 A0A0P5JZ33 A0A0P6HEP5 A0A067QI31 A0A0P5K061 A0A0P5LPF8 A0A0P5SEQ5 A0A0P5LD82 A0A0P5VUB4 A0A0P5JBM5 A0A0P5L5V1 A0A0P6DB64 A0A0P5KKB0 A0A0N7ZUZ1 A0A0P5PC38 A0A0P5SY78 A0A0P6CYU6 A0A0P5N754 A0A0P6E3W7 A0A0P5KYN3 A0A0N8BZ42 A0A0P5KRY9 A0A0P6DHG3 A0A0P5L176 A0A0P5LJY0 A0A0P5MXZ2 A0A0P5LIH6 A0A0P5JVC6 A0A0P5P1H8 A0A0P5NG43 A0A0N8DVZ9 A0A0P6DW90 A0A0P5QRW2 A0A0P5VZK7 A0A0P5W213 A0A0P4Z5F4 A0A0P5Q110 A0A0P5UHB8 A0A0P4ZW92 A0A0N8AVA9 A0A2J7Q872 A0A0P5ID55 A0A0P5ZET5 A0A0P6ERF7 A0A0P5JJ67 A0A0P5BNP1 A0A0P5X2K1 A0A0P5BR09 A0A0P5VZH1 A0A0P4ZVU9 A0A2J7Q871 A0A0P5Q230 A0A0P5WVY8 A0A0N8AES1 A0A0P5PMB6 A0A0P5M0M0
Pubmed
EMBL
NWSH01000735
PCG74506.1
ODYU01011946
SOQ57983.1
KQ461103
KPJ09368.1
+ More
KQ459589 KPI97579.1 QOIP01000006 RLU21133.1 KK107020 EZA62576.1 KQ977106 KYN05755.1 ADTU01027574 KQ982790 KYQ50675.1 NEVH01016973 PNF24782.1 PNF24781.1 KQ976730 KYM76400.1 KQ981993 KYN31125.1 AAZX01003980 KQ435821 KOX72398.1 GL451420 EFN79325.1 KQ769068 OAD52987.1 KQ971344 KYB27089.1 KQ414727 KOC62505.1 GDIP01083235 JAM20480.1 PNF24779.1 GDIQ01230930 JAK20795.1 GDIP01083234 JAM20481.1 GDIP01060631 JAM43084.1 GDIP01192963 JAJ30439.1 GDIQ01106169 JAL45557.1 GDIP01198071 JAJ25331.1 GDIP01207108 JAJ16294.1 GDIQ01072335 JAN22402.1 GDIQ01213933 JAK37792.1 GDIQ01072334 JAN22403.1 GDIQ01199533 JAK52192.1 KQ434819 KZC06953.1 GDIQ01213934 JAK37791.1 GDIQ01128517 JAL23209.1 GDIQ01180349 GDIQ01113569 GDIQ01047395 GDIQ01047394 JAN47342.1 GDIQ01180348 JAK71377.1 GL732539 EFX82660.1 GDIQ01110471 JAL41255.1 GDIQ01106170 JAL45556.1 GDIQ01234130 JAK17595.1 GDIQ01198193 GDIQ01159505 GDIQ01157603 GDIQ01152555 GDIQ01141209 GDIQ01139009 GDIQ01136517 GDIQ01126621 GDIQ01112169 GDIQ01110470 GDIQ01106171 GDIQ01094923 GDIQ01045549 GDIQ01043393 JAK94122.1 GDIQ01126620 JAL25106.1 GDIQ01193089 JAK58636.1 GDIQ01194890 GDIQ01141210 GDIQ01124918 GDIQ01096777 GDIQ01094924 GDIQ01043394 JAN51343.1 KK853332 KDR08398.1 GDIQ01196634 JAK55091.1 GDIQ01175545 JAK76180.1 GDIQ01096776 JAL54950.1 GDIQ01196633 JAK55092.1 GDIP01109732 GDIP01109731 GDIP01094486 JAL93982.1 GDIQ01201942 GDIQ01130725 JAK49783.1 GDIQ01199532 JAK52193.1 GDIQ01081577 JAN13160.1 GDIQ01184040 GDIQ01184038 JAK67687.1 GDIP01210009 JAJ13393.1 GDIQ01201944 GDIQ01142905 GDIQ01130726 JAL21000.1 GDIP01138374 JAL65340.1 GDIQ01086478 JAN08259.1 GDIQ01218041 GDIQ01216926 GDIQ01159506 GDIQ01157604 GDIQ01112170 GDIQ01083431 JAK94121.1 GDIQ01068443 JAN26294.1 GDIQ01177390 JAK74335.1 GDIQ01201945 GDIQ01185844 GDIQ01130723 GDIQ01129823 GDIQ01086479 GDIQ01068444 JAL21003.1 GDIQ01187394 JAK64331.1 GDIQ01187393 GDIQ01086475 JAN08262.1 GDIQ01184039 JAK67686.1 GDIQ01177391 GDIQ01168549 GDIQ01032453 JAK83176.1 GDIQ01161050 JAK90675.1 GDIQ01177392 JAK74333.1 GDIQ01193088 JAK58637.1 GDIQ01136518 JAL15208.1 GDIQ01142906 JAL08820.1 GDIQ01201943 GDIQ01101079 GDIQ01086476 JAN08261.1 GDIQ01086477 JAN08260.1 GDIQ01130722 JAL21004.1 GDIP01092977 JAM10738.1 GDIP01092978 JAM10737.1 GDIP01217663 GDIP01215318 GDIP01198911 GDIP01198070 JAJ05739.1 GDIQ01130724 JAL21002.1 GDIP01130121 JAL73593.1 GDIP01207167 JAJ16235.1 GDIQ01239387 GDIQ01239386 JAK12338.1 PNF24783.1 GDIQ01239385 JAK12340.1 GDIP01045198 JAM58517.1 GDIQ01072333 JAN22404.1 GDIQ01198194 JAK53531.1 GDIP01186966 GDIP01186965 JAJ36436.1 GDIP01078776 JAM24939.1 GDIP01185823 JAJ37579.1 GDIP01093025 JAM10690.1 GDIP01207166 JAJ16236.1 PNF24778.1 GDIQ01120212 JAL31514.1 GDIP01081277 JAM22438.1 GDIP01154569 JAJ68833.1 GDIQ01131973 JAL19753.1 GDIQ01165208 JAK86517.1
KQ459589 KPI97579.1 QOIP01000006 RLU21133.1 KK107020 EZA62576.1 KQ977106 KYN05755.1 ADTU01027574 KQ982790 KYQ50675.1 NEVH01016973 PNF24782.1 PNF24781.1 KQ976730 KYM76400.1 KQ981993 KYN31125.1 AAZX01003980 KQ435821 KOX72398.1 GL451420 EFN79325.1 KQ769068 OAD52987.1 KQ971344 KYB27089.1 KQ414727 KOC62505.1 GDIP01083235 JAM20480.1 PNF24779.1 GDIQ01230930 JAK20795.1 GDIP01083234 JAM20481.1 GDIP01060631 JAM43084.1 GDIP01192963 JAJ30439.1 GDIQ01106169 JAL45557.1 GDIP01198071 JAJ25331.1 GDIP01207108 JAJ16294.1 GDIQ01072335 JAN22402.1 GDIQ01213933 JAK37792.1 GDIQ01072334 JAN22403.1 GDIQ01199533 JAK52192.1 KQ434819 KZC06953.1 GDIQ01213934 JAK37791.1 GDIQ01128517 JAL23209.1 GDIQ01180349 GDIQ01113569 GDIQ01047395 GDIQ01047394 JAN47342.1 GDIQ01180348 JAK71377.1 GL732539 EFX82660.1 GDIQ01110471 JAL41255.1 GDIQ01106170 JAL45556.1 GDIQ01234130 JAK17595.1 GDIQ01198193 GDIQ01159505 GDIQ01157603 GDIQ01152555 GDIQ01141209 GDIQ01139009 GDIQ01136517 GDIQ01126621 GDIQ01112169 GDIQ01110470 GDIQ01106171 GDIQ01094923 GDIQ01045549 GDIQ01043393 JAK94122.1 GDIQ01126620 JAL25106.1 GDIQ01193089 JAK58636.1 GDIQ01194890 GDIQ01141210 GDIQ01124918 GDIQ01096777 GDIQ01094924 GDIQ01043394 JAN51343.1 KK853332 KDR08398.1 GDIQ01196634 JAK55091.1 GDIQ01175545 JAK76180.1 GDIQ01096776 JAL54950.1 GDIQ01196633 JAK55092.1 GDIP01109732 GDIP01109731 GDIP01094486 JAL93982.1 GDIQ01201942 GDIQ01130725 JAK49783.1 GDIQ01199532 JAK52193.1 GDIQ01081577 JAN13160.1 GDIQ01184040 GDIQ01184038 JAK67687.1 GDIP01210009 JAJ13393.1 GDIQ01201944 GDIQ01142905 GDIQ01130726 JAL21000.1 GDIP01138374 JAL65340.1 GDIQ01086478 JAN08259.1 GDIQ01218041 GDIQ01216926 GDIQ01159506 GDIQ01157604 GDIQ01112170 GDIQ01083431 JAK94121.1 GDIQ01068443 JAN26294.1 GDIQ01177390 JAK74335.1 GDIQ01201945 GDIQ01185844 GDIQ01130723 GDIQ01129823 GDIQ01086479 GDIQ01068444 JAL21003.1 GDIQ01187394 JAK64331.1 GDIQ01187393 GDIQ01086475 JAN08262.1 GDIQ01184039 JAK67686.1 GDIQ01177391 GDIQ01168549 GDIQ01032453 JAK83176.1 GDIQ01161050 JAK90675.1 GDIQ01177392 JAK74333.1 GDIQ01193088 JAK58637.1 GDIQ01136518 JAL15208.1 GDIQ01142906 JAL08820.1 GDIQ01201943 GDIQ01101079 GDIQ01086476 JAN08261.1 GDIQ01086477 JAN08260.1 GDIQ01130722 JAL21004.1 GDIP01092977 JAM10738.1 GDIP01092978 JAM10737.1 GDIP01217663 GDIP01215318 GDIP01198911 GDIP01198070 JAJ05739.1 GDIQ01130724 JAL21002.1 GDIP01130121 JAL73593.1 GDIP01207167 JAJ16235.1 GDIQ01239387 GDIQ01239386 JAK12338.1 PNF24783.1 GDIQ01239385 JAK12340.1 GDIP01045198 JAM58517.1 GDIQ01072333 JAN22404.1 GDIQ01198194 JAK53531.1 GDIP01186966 GDIP01186965 JAJ36436.1 GDIP01078776 JAM24939.1 GDIP01185823 JAJ37579.1 GDIP01093025 JAM10690.1 GDIP01207166 JAJ16236.1 PNF24778.1 GDIQ01120212 JAL31514.1 GDIP01081277 JAM22438.1 GDIP01154569 JAJ68833.1 GDIQ01131973 JAL19753.1 GDIQ01165208 JAK86517.1
Proteomes
Interpro
SUPFAM
SSF52009
SSF52009
Gene 3D
ProteinModelPortal
A0A2A4JRX2
A0A2H1WY32
A0A194QV09
A0A194PXV0
A0A3L8DLP2
A0A026X3J0
+ More
A0A151ILI8 A0A158NVZ5 A0A151WSS2 A0A2J7Q877 A0A2J7Q868 A0A151HY46 A0A195ES97 K7JPF5 A0A0N0BF09 E2BY46 A0A310S601 A0A139WGR7 A0A0L7QV50 A0A0P5WPZ2 A0A2J7Q876 A0A0P5H5T3 A0A0P5WRL6 A0A0N8D7Q2 A0A0P5AWX9 A0A0P5RGS6 A0A0P5ATU0 A0A0P5A875 A0A0P6DU69 A0A0P5IFV5 A0A0P6DYB3 A0A0P5JS45 A0A154P524 A0A0P5IDP6 A0A0P5PYV7 A0A0P6GB77 A0A0P5KRB8 E9GD24 A0A0P5S2P4 A0A0P5R3H5 A0A0P5GYA6 A0A0N8BPI2 A0A0P5PLP8 A0A0P5JZ33 A0A0P6HEP5 A0A067QI31 A0A0P5K061 A0A0P5LPF8 A0A0P5SEQ5 A0A0P5LD82 A0A0P5VUB4 A0A0P5JBM5 A0A0P5L5V1 A0A0P6DB64 A0A0P5KKB0 A0A0N7ZUZ1 A0A0P5PC38 A0A0P5SY78 A0A0P6CYU6 A0A0P5N754 A0A0P6E3W7 A0A0P5KYN3 A0A0N8BZ42 A0A0P5KRY9 A0A0P6DHG3 A0A0P5L176 A0A0P5LJY0 A0A0P5MXZ2 A0A0P5LIH6 A0A0P5JVC6 A0A0P5P1H8 A0A0P5NG43 A0A0N8DVZ9 A0A0P6DW90 A0A0P5QRW2 A0A0P5VZK7 A0A0P5W213 A0A0P4Z5F4 A0A0P5Q110 A0A0P5UHB8 A0A0P4ZW92 A0A0N8AVA9 A0A2J7Q872 A0A0P5ID55 A0A0P5ZET5 A0A0P6ERF7 A0A0P5JJ67 A0A0P5BNP1 A0A0P5X2K1 A0A0P5BR09 A0A0P5VZH1 A0A0P4ZVU9 A0A2J7Q871 A0A0P5Q230 A0A0P5WVY8 A0A0N8AES1 A0A0P5PMB6 A0A0P5M0M0
A0A151ILI8 A0A158NVZ5 A0A151WSS2 A0A2J7Q877 A0A2J7Q868 A0A151HY46 A0A195ES97 K7JPF5 A0A0N0BF09 E2BY46 A0A310S601 A0A139WGR7 A0A0L7QV50 A0A0P5WPZ2 A0A2J7Q876 A0A0P5H5T3 A0A0P5WRL6 A0A0N8D7Q2 A0A0P5AWX9 A0A0P5RGS6 A0A0P5ATU0 A0A0P5A875 A0A0P6DU69 A0A0P5IFV5 A0A0P6DYB3 A0A0P5JS45 A0A154P524 A0A0P5IDP6 A0A0P5PYV7 A0A0P6GB77 A0A0P5KRB8 E9GD24 A0A0P5S2P4 A0A0P5R3H5 A0A0P5GYA6 A0A0N8BPI2 A0A0P5PLP8 A0A0P5JZ33 A0A0P6HEP5 A0A067QI31 A0A0P5K061 A0A0P5LPF8 A0A0P5SEQ5 A0A0P5LD82 A0A0P5VUB4 A0A0P5JBM5 A0A0P5L5V1 A0A0P6DB64 A0A0P5KKB0 A0A0N7ZUZ1 A0A0P5PC38 A0A0P5SY78 A0A0P6CYU6 A0A0P5N754 A0A0P6E3W7 A0A0P5KYN3 A0A0N8BZ42 A0A0P5KRY9 A0A0P6DHG3 A0A0P5L176 A0A0P5LJY0 A0A0P5MXZ2 A0A0P5LIH6 A0A0P5JVC6 A0A0P5P1H8 A0A0P5NG43 A0A0N8DVZ9 A0A0P6DW90 A0A0P5QRW2 A0A0P5VZK7 A0A0P5W213 A0A0P4Z5F4 A0A0P5Q110 A0A0P5UHB8 A0A0P4ZW92 A0A0N8AVA9 A0A2J7Q872 A0A0P5ID55 A0A0P5ZET5 A0A0P6ERF7 A0A0P5JJ67 A0A0P5BNP1 A0A0P5X2K1 A0A0P5BR09 A0A0P5VZH1 A0A0P4ZVU9 A0A2J7Q871 A0A0P5Q230 A0A0P5WVY8 A0A0N8AES1 A0A0P5PMB6 A0A0P5M0M0
PDB
5HV3
E-value=1.52775e-35,
Score=379
Ontologies
GO
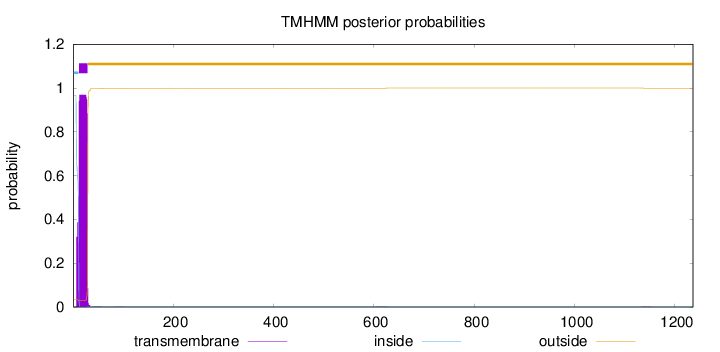
Topology
Length:
1236
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.4226199999999
Exp number, first 60 AAs:
19.38928
Total prob of N-in:
0.96602
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 29
outside
30 - 1236
Population Genetic Test Statistics
Pi
41.001605
Theta
20.225573
Tajima's D
-1.073885
CLR
304.105919
CSRT
0.122993850307485
Interpretation
Uncertain
