Pre Gene Modal
BGIBMGA002597
Annotation
putative_tyrosine-protein_kinase_drl_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.376
Sequence
CDS
ATGCACCCACCGACCTTTAACATTTCGAACGAGGGCTACGTGCCGACCGAACCGCAGACGTGGCGAGTGGACTTGCCATGTATGCGGACGATCGCCGCTGAAGTTGATCTCACTATATCACTCAATGTTTCGACCGGCTCGTCATCTACGACTTTGCATTTTCGACGGAAGAAGATATGTTTGAAAGAGGCCATCCGACCTGCCGTTAGAGTAGATAGCGTGCCGCACGCGCCTACCTCAGCGGATATTTTCTACGCTGGATCCGGATGCGCTGGAGGCATTATACTAATAGCTGCAATTGCCGCGGCGGCTTGCAGAGCACGAGCCAGAAAACTGAGACGGGCTCGCAGCGATGAACAGGACGCTCACGCGTTCTTACCAGACTCGTCGTGCAAGTCGGCCGCCAGCTATACCAGTTACAGGAGACCCACTTCGCTTCCAGCCGTAGCTGCTGAAGAACGAGCCAGAGATCTACAGCAGAGGATAGCTGAGTTAACAATTCAACGGTGTCGAGTGAGGTTGCGTTCCGTAGCGATGGAGGGGACGTTCGGCCGAGTCTACCGCGGGACTTATGCTGATGAGGAAGCACGAGAGCAAGAAGTTCTCGTAAAGACCGTCGCCGAACATGCATCTCAAGTACAGGTATCGCTGCTTCTTCAAGAAGGCTGCATGTTGTACGGACTCCATCACGAACGTGTGCTGTCCGTTCTCGGAGTTAGCATTGAGGACCAAACGGCTCCGTTCTTGTTGTATCCTTGGGATGCTGGATGGAGAAACATGAAGCTATTCTTGCTGGCGTGCAGAGGAGTGACCATTGGAGGGGCACCCGGGGGAGCACCTCCGCCGCCTCTAACGACTCAGCACGTCGTCAGGATGGCGCTACATGCCCTCGATGGCTTGCTCTATCTTCACTCTCAACACGTGCTACATAAAGACATCGCCGCTAGGAACTGCATAGTGGATGAAAATCTGCGAGTGATGATAGCCGACAACGCGTTATCGAGGGATCTCTTCCCGGCAGACTACCATTGCTTAGGGGATAATGAGAACCGGCCTATCAAATGGCTGGCGTTAGAAGCTTTGACGAAAAGACAGTTCAGCCCGGCGGCGGACGTCTGGGCTTTAGGGGTGCTTCTGTGGGAGTTAACAACGTTGGCTCACCAACCTTACGCGGAGGTCGATCCCTTTGAGGTGGCTGCGTATTTAAGAGACGGATACCGGTTGCAACAGCCGGCGAACTGTCCTGATGAATTGTTTGCTGTGATGGCGTACTGCTGGGCAATGTCACCAGACGACCGGCCCACTCTCCCTCAGCTGCAGATCTTCCTTCGAGACTTCCACACGCAATTAACGAGATTCGTTTAG
Protein
MHPPTFNISNEGYVPTEPQTWRVDLPCMRTIAAEVDLTISLNVSTGSSSTTLHFRRKKICLKEAIRPAVRVDSVPHAPTSADIFYAGSGCAGGIILIAAIAAAACRARARKLRRARSDEQDAHAFLPDSSCKSAASYTSYRRPTSLPAVAAEERARDLQQRIAELTIQRCRVRLRSVAMEGTFGRVYRGTYADEEAREQEVLVKTVAEHASQVQVSLLLQEGCMLYGLHHERVLSVLGVSIEDQTAPFLLYPWDAGWRNMKLFLLACRGVTIGGAPGGAPPPPLTTQHVVRMALHALDGLLYLHSQHVLHKDIAARNCIVDENLRVMIADNALSRDLFPADYHCLGDNENRPIKWLALEALTKRQFSPAADVWALGVLLWELTTLAHQPYAEVDPFEVAAYLRDGYRLQQPANCPDELFAVMAYCWAMSPDDRPTLPQLQIFLRDFHTQLTRFV
Summary
Similarity
Belongs to the calycin superfamily. Lipocalin family.
Uniprot
H9IZB2
A0A212ES33
A0A2A4JGY0
A0A194R0M0
A0A0L7LS08
A0A194Q2G5
+ More
A0A2J7QF99 A0A0T6AUW0 A0A0U3TKW7 D6W7I8 E0W415 A0A1Y1LXG5 A0A1W4WP05 A0A1W4WYR6 U4TSR8 A0A026WYZ9 E2ABN1 A0A3L8D3R9 A0A0C9RJZ1 A0A1B6DI20 A0A195C8Y1 E9INZ5 E2BV04 F4WGZ4 A0A088AHA8 A0A154P1L5 A0A2A3E8A5 A0A158NVL2 A0A195E3X7 A0A151X5B3 A0A195BUU5 A0A151K1G5 A0A1B6LZC5 A0A0L7QP32 A0A1B6GG95 A0A3Q0J0S9 A0A0P6IW15 A0A1J1IWS7 A0A0A1XGL9 A0A1B0DIA8 A0A1B0GQB0 A0A1I8MZV1 J9JT54 Q17PG3 A0A182NTN9 A0A084WD96 A0A182SH29 A0A182W7D5 A0A182YSD3 Q7PND0 A0A182L1U5 A0A182HUZ6 A0A182RWI4 A0A182MC97 A0A182VM15 A0A1S4GWY8 A0A182QAV6 A0A084WD97 A0A2M4DNV6 A0A0K8UE71 A0A182TYH8 A0A182JHK3 A0A2H8TJ84 A0A0K8TWZ7 A0A2S2P021 A0A084WCL9 A0A1A9WHI0 A0A1B0AH32 A0A1B0GAD2 A0A1A9XRN5 A0A1B0BET8 A0A1A9UEV5 A0A1I8P122 A0A3P8TU24 B4JP95 B4M951 B4MTH9 B4KKE7 A0A3B4VIS2 A0A3B5LAG8 Q29K37 B4GWU5
A0A2J7QF99 A0A0T6AUW0 A0A0U3TKW7 D6W7I8 E0W415 A0A1Y1LXG5 A0A1W4WP05 A0A1W4WYR6 U4TSR8 A0A026WYZ9 E2ABN1 A0A3L8D3R9 A0A0C9RJZ1 A0A1B6DI20 A0A195C8Y1 E9INZ5 E2BV04 F4WGZ4 A0A088AHA8 A0A154P1L5 A0A2A3E8A5 A0A158NVL2 A0A195E3X7 A0A151X5B3 A0A195BUU5 A0A151K1G5 A0A1B6LZC5 A0A0L7QP32 A0A1B6GG95 A0A3Q0J0S9 A0A0P6IW15 A0A1J1IWS7 A0A0A1XGL9 A0A1B0DIA8 A0A1B0GQB0 A0A1I8MZV1 J9JT54 Q17PG3 A0A182NTN9 A0A084WD96 A0A182SH29 A0A182W7D5 A0A182YSD3 Q7PND0 A0A182L1U5 A0A182HUZ6 A0A182RWI4 A0A182MC97 A0A182VM15 A0A1S4GWY8 A0A182QAV6 A0A084WD97 A0A2M4DNV6 A0A0K8UE71 A0A182TYH8 A0A182JHK3 A0A2H8TJ84 A0A0K8TWZ7 A0A2S2P021 A0A084WCL9 A0A1A9WHI0 A0A1B0AH32 A0A1B0GAD2 A0A1A9XRN5 A0A1B0BET8 A0A1A9UEV5 A0A1I8P122 A0A3P8TU24 B4JP95 B4M951 B4MTH9 B4KKE7 A0A3B4VIS2 A0A3B5LAG8 Q29K37 B4GWU5
Pubmed
EMBL
BABH01014364
AGBW02012871
OWR44302.1
NWSH01001502
PCG71036.1
KQ461103
+ More
KPJ09386.1 JTDY01000231 KOB78164.1 KQ459589 KPI97595.1 NEVH01015301 PNF27271.1 LJIG01022756 KRT78888.1 KT355579 ALV82498.1 KQ971307 EFA11279.1 DS235886 EEB20371.1 GEZM01049965 JAV75727.1 KB631602 ERL84559.1 KK107061 EZA61280.1 GL438334 EFN69133.1 QOIP01000014 RLU15040.1 GBYB01008515 JAG78282.1 GEDC01011965 JAS25333.1 KQ978072 KYM97287.1 GL764397 EFZ17775.1 GL450776 EFN80484.1 GL888148 EGI66473.1 KQ434786 KZC05234.1 KZ288347 PBC27512.1 ADTU01027236 ADTU01027237 ADTU01027238 ADTU01027239 KQ979701 KYN19617.1 KQ982515 KYQ55595.1 KQ976409 KYM90966.1 KQ981248 KYN44243.1 GEBQ01010993 JAT28984.1 KQ414836 KOC60383.1 GECZ01008305 JAS61464.1 GDUN01000761 JAN95158.1 CVRI01000059 CRL03614.1 GBXI01004644 JAD09648.1 AJVK01034329 AJVK01036058 ABLF02026171 CH477191 EAT48654.1 ATLV01022956 KE525338 KFB48190.1 AAAB01008964 EAA12391.5 APCN01002487 AXCM01017601 AXCN02000795 ATLV01022969 KFB48191.1 GGFL01014620 MBW78798.1 GDHF01027370 GDHF01013735 JAI24944.1 JAI38579.1 GFXV01001947 MBW13752.1 GDHF01033709 GDHF01028451 JAI18605.1 JAI23863.1 GGMR01010166 MBY22785.1 ATLV01022743 KE525336 KFB47963.1 CCAG010022674 JXJN01013014 CH916372 EDV98725.1 CH940654 EDW57727.2 CH963852 EDW75418.2 CH933807 EDW11597.2 CH379061 EAL33341.2 CH479195 EDW27272.1
KPJ09386.1 JTDY01000231 KOB78164.1 KQ459589 KPI97595.1 NEVH01015301 PNF27271.1 LJIG01022756 KRT78888.1 KT355579 ALV82498.1 KQ971307 EFA11279.1 DS235886 EEB20371.1 GEZM01049965 JAV75727.1 KB631602 ERL84559.1 KK107061 EZA61280.1 GL438334 EFN69133.1 QOIP01000014 RLU15040.1 GBYB01008515 JAG78282.1 GEDC01011965 JAS25333.1 KQ978072 KYM97287.1 GL764397 EFZ17775.1 GL450776 EFN80484.1 GL888148 EGI66473.1 KQ434786 KZC05234.1 KZ288347 PBC27512.1 ADTU01027236 ADTU01027237 ADTU01027238 ADTU01027239 KQ979701 KYN19617.1 KQ982515 KYQ55595.1 KQ976409 KYM90966.1 KQ981248 KYN44243.1 GEBQ01010993 JAT28984.1 KQ414836 KOC60383.1 GECZ01008305 JAS61464.1 GDUN01000761 JAN95158.1 CVRI01000059 CRL03614.1 GBXI01004644 JAD09648.1 AJVK01034329 AJVK01036058 ABLF02026171 CH477191 EAT48654.1 ATLV01022956 KE525338 KFB48190.1 AAAB01008964 EAA12391.5 APCN01002487 AXCM01017601 AXCN02000795 ATLV01022969 KFB48191.1 GGFL01014620 MBW78798.1 GDHF01027370 GDHF01013735 JAI24944.1 JAI38579.1 GFXV01001947 MBW13752.1 GDHF01033709 GDHF01028451 JAI18605.1 JAI23863.1 GGMR01010166 MBY22785.1 ATLV01022743 KE525336 KFB47963.1 CCAG010022674 JXJN01013014 CH916372 EDV98725.1 CH940654 EDW57727.2 CH963852 EDW75418.2 CH933807 EDW11597.2 CH379061 EAL33341.2 CH479195 EDW27272.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000037510
UP000053268
+ More
UP000235965 UP000007266 UP000009046 UP000192223 UP000030742 UP000053097 UP000000311 UP000279307 UP000078542 UP000008237 UP000007755 UP000005203 UP000076502 UP000242457 UP000005205 UP000078492 UP000075809 UP000078540 UP000078541 UP000053825 UP000079169 UP000183832 UP000092462 UP000095301 UP000007819 UP000008820 UP000075884 UP000030765 UP000075901 UP000075920 UP000076408 UP000007062 UP000075882 UP000075840 UP000075900 UP000075883 UP000075903 UP000075886 UP000075902 UP000075880 UP000091820 UP000092445 UP000092444 UP000092443 UP000092460 UP000078200 UP000095300 UP000265080 UP000001070 UP000008792 UP000007798 UP000009192 UP000261420 UP000261380 UP000001819 UP000008744
UP000235965 UP000007266 UP000009046 UP000192223 UP000030742 UP000053097 UP000000311 UP000279307 UP000078542 UP000008237 UP000007755 UP000005203 UP000076502 UP000242457 UP000005205 UP000078492 UP000075809 UP000078540 UP000078541 UP000053825 UP000079169 UP000183832 UP000092462 UP000095301 UP000007819 UP000008820 UP000075884 UP000030765 UP000075901 UP000075920 UP000076408 UP000007062 UP000075882 UP000075840 UP000075900 UP000075883 UP000075903 UP000075886 UP000075902 UP000075880 UP000091820 UP000092445 UP000092444 UP000092443 UP000092460 UP000078200 UP000095300 UP000265080 UP000001070 UP000008792 UP000007798 UP000009192 UP000261420 UP000261380 UP000001819 UP000008744
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IZB2
A0A212ES33
A0A2A4JGY0
A0A194R0M0
A0A0L7LS08
A0A194Q2G5
+ More
A0A2J7QF99 A0A0T6AUW0 A0A0U3TKW7 D6W7I8 E0W415 A0A1Y1LXG5 A0A1W4WP05 A0A1W4WYR6 U4TSR8 A0A026WYZ9 E2ABN1 A0A3L8D3R9 A0A0C9RJZ1 A0A1B6DI20 A0A195C8Y1 E9INZ5 E2BV04 F4WGZ4 A0A088AHA8 A0A154P1L5 A0A2A3E8A5 A0A158NVL2 A0A195E3X7 A0A151X5B3 A0A195BUU5 A0A151K1G5 A0A1B6LZC5 A0A0L7QP32 A0A1B6GG95 A0A3Q0J0S9 A0A0P6IW15 A0A1J1IWS7 A0A0A1XGL9 A0A1B0DIA8 A0A1B0GQB0 A0A1I8MZV1 J9JT54 Q17PG3 A0A182NTN9 A0A084WD96 A0A182SH29 A0A182W7D5 A0A182YSD3 Q7PND0 A0A182L1U5 A0A182HUZ6 A0A182RWI4 A0A182MC97 A0A182VM15 A0A1S4GWY8 A0A182QAV6 A0A084WD97 A0A2M4DNV6 A0A0K8UE71 A0A182TYH8 A0A182JHK3 A0A2H8TJ84 A0A0K8TWZ7 A0A2S2P021 A0A084WCL9 A0A1A9WHI0 A0A1B0AH32 A0A1B0GAD2 A0A1A9XRN5 A0A1B0BET8 A0A1A9UEV5 A0A1I8P122 A0A3P8TU24 B4JP95 B4M951 B4MTH9 B4KKE7 A0A3B4VIS2 A0A3B5LAG8 Q29K37 B4GWU5
A0A2J7QF99 A0A0T6AUW0 A0A0U3TKW7 D6W7I8 E0W415 A0A1Y1LXG5 A0A1W4WP05 A0A1W4WYR6 U4TSR8 A0A026WYZ9 E2ABN1 A0A3L8D3R9 A0A0C9RJZ1 A0A1B6DI20 A0A195C8Y1 E9INZ5 E2BV04 F4WGZ4 A0A088AHA8 A0A154P1L5 A0A2A3E8A5 A0A158NVL2 A0A195E3X7 A0A151X5B3 A0A195BUU5 A0A151K1G5 A0A1B6LZC5 A0A0L7QP32 A0A1B6GG95 A0A3Q0J0S9 A0A0P6IW15 A0A1J1IWS7 A0A0A1XGL9 A0A1B0DIA8 A0A1B0GQB0 A0A1I8MZV1 J9JT54 Q17PG3 A0A182NTN9 A0A084WD96 A0A182SH29 A0A182W7D5 A0A182YSD3 Q7PND0 A0A182L1U5 A0A182HUZ6 A0A182RWI4 A0A182MC97 A0A182VM15 A0A1S4GWY8 A0A182QAV6 A0A084WD97 A0A2M4DNV6 A0A0K8UE71 A0A182TYH8 A0A182JHK3 A0A2H8TJ84 A0A0K8TWZ7 A0A2S2P021 A0A084WCL9 A0A1A9WHI0 A0A1B0AH32 A0A1B0GAD2 A0A1A9XRN5 A0A1B0BET8 A0A1A9UEV5 A0A1I8P122 A0A3P8TU24 B4JP95 B4M951 B4MTH9 B4KKE7 A0A3B4VIS2 A0A3B5LAG8 Q29K37 B4GWU5
PDB
5DG5
E-value=4.2659e-43,
Score=440
Ontologies
GO
GO:0005524
GO:0016021
GO:0004672
GO:0004713
GO:0007409
GO:0018108
GO:0017147
GO:0005887
GO:0043235
GO:0007399
GO:0007169
GO:0016055
GO:0004714
GO:0016301
GO:0007165
GO:0007435
GO:0016204
GO:0046982
GO:0007411
GO:0007223
GO:0060030
GO:0060028
GO:0051209
GO:0005886
GO:0006468
GO:0005085
GO:0016776
GO:0006172
GO:0006631
GO:0006635
GO:0006281
GO:0048384
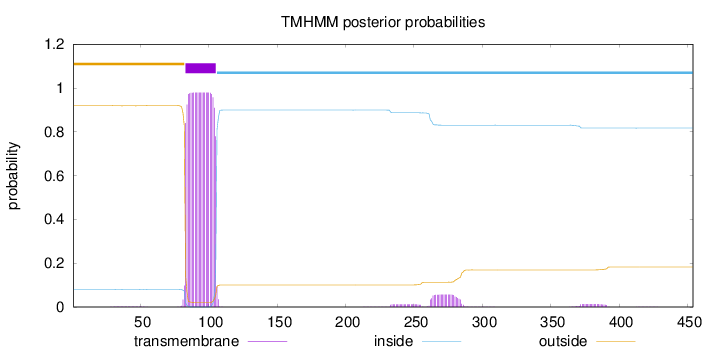
Topology
Length:
454
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.10698
Exp number, first 60 AAs:
0.04284
Total prob of N-in:
0.08014
outside
1 - 82
TMhelix
83 - 105
inside
106 - 454
Population Genetic Test Statistics
Pi
212.562894
Theta
181.672856
Tajima's D
0.606722
CLR
0.48093
CSRT
0.542922853857307
Interpretation
Uncertain
