Gene
KWMTBOMO02533
Pre Gene Modal
BGIBMGA002692
Annotation
PREDICTED:_guanine_nucleotide_exchange_factor_DBS_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.495 Nuclear Reliability : 1.487
Sequence
CDS
ATGAACTCCCCTCCACGGTGCGCTTCGGAGCGCTATGCATGTCCTTCTTCAAACGAAGCTTGCGGAGAGTACTTAATGGTAGGCCGCGGCTTGGCTCTGCTCCTGGCATTGCTGACGTCCATGAGCGACGGTGACCACTTACCATCAGAAGCGGAGTGTGATGGGTGCGGCAGCGAAGGTGACGAAGCTCGCAATGTGGGAGAGCTTTTACTGCCACAATACGCGCTGGCCACCGGAGGACTGGCCAAAGACCACCGTCCCCTCATCACCTTCCCGGACAACAATAACTTTCATCTCATCACTGCCTCCGAGTACCGAAGACTGTTGCTCTACCTCACCTCTGTGCCTTCGATACTAGAAGCAGATATGGGCTTCCACATTATAGTGGACAGAAGAAAGGACAGATGGAATTCTGTAAAAACAGTATTACTTAGGATCTCAGAATTCTTCCCTGGTATAATCCACGCGGTTTACGTTCTTCGCCCGTCAAGTTTCCTGCAGAAAGCGCTGTCCGAGGTCAGCAGCAAACTGTTTAAGGAGGAATTCAGGTTCAAAGTGCTAGTGTGCTCCACGGTCGATGAACTCTACGAGCACTTCGAAAGGTCTCAGCTGACTCCTGACCTTGGCGGGGAGTTACAATATTCACATTCTGAATGGATACAGCAGCGAATAGCTTTAGAGAAATTTTCAACGCTTATGAAAGAGACATCGACGAAGCTGGACGATTTCATGCACGAGATAGTCGACTGCGATATGGGAAACGATCCCACACAAACCAAAGAATTATTAGACTGTCAGGGAACTAGGTATAAAGCTTTAAAATCTGAATTATCAATAGCAACAACACAAGGAGAAGATTTATTAACACAAGTGCGGAAGCCGAATCTTACGTACAACATTATAAGTCATGTAGCTGCCGTCGAAAGGTTACTAGTACAACTGGAAGAGACAGAGAAGCAGTTTGATAATTTCTGGCAGAAGCATTCCACAAAACTCAATCACTGGTTGAAGTTTAGAACGTTTTTATTGAATTTCAAACAGATGCAGGCAACTTTAGATCATCATCTAAAAGCAGCGTGTGACATGACCGAAGTGGGAGAAACAGCCAGTAGAGTGGAAAGTCTCATTCAGGAAGCGCACAACTTTGAGAAATTATGCAACTGTGATCTTGATACTGCATCTTCAGTGATCGATGATGGTGATAAACTAATGCAAGATCCGCTAAGTTCCGTCGATCATATAGAATCAAAATGCGAAGAACTAAGACGTACAAGCGCCCTGCTTATCGACAAGATACAAAAACGAAATTTACTACTCACGAAAGCTAGAGAACTAATGGATAGGATAGATAAGGCGAACGAATGGTGTGCAACTGGAGTAGAGCTGTTAGCCGGAGAAGGAGGTCTGCTGGCTGTAGACAAGCTGCTGGAAGACGCTCAGTCCTTTGGGCTGACAGCACCTGAACAGTTCCGAGACATGCTTATGCAGTCCGCCACCCAAGAAACAAGAGCTTTGGTCACTCAAGTAGCACAAAGAGTTGAGGACGTATGGTTGATGGTGTCAGTGAAACGAGCTTCACTGCAACGCGCGGCCGCCAGACCGGCTCGCCCCGTACAGTCCGTGCCCCCGCAGAGCGCTGCCCTACCAACTCCGGCTAATCAACAGATGCAAATCCGTTAA
Protein
MNSPPRCASERYACPSSNEACGEYLMVGRGLALLLALLTSMSDGDHLPSEAECDGCGSEGDEARNVGELLLPQYALATGGLAKDHRPLITFPDNNNFHLITASEYRRLLLYLTSVPSILEADMGFHIIVDRRKDRWNSVKTVLLRISEFFPGIIHAVYVLRPSSFLQKALSEVSSKLFKEEFRFKVLVCSTVDELYEHFERSQLTPDLGGELQYSHSEWIQQRIALEKFSTLMKETSTKLDDFMHEIVDCDMGNDPTQTKELLDCQGTRYKALKSELSIATTQGEDLLTQVRKPNLTYNIISHVAAVERLLVQLEETEKQFDNFWQKHSTKLNHWLKFRTFLLNFKQMQATLDHHLKAACDMTEVGETASRVESLIQEAHNFEKLCNCDLDTASSVIDDGDKLMQDPLSSVDHIESKCEELRRTSALLIDKIQKRNLLLTKARELMDRIDKANEWCATGVELLAGEGGLLAVDKLLEDAQSFGLTAPEQFRDMLMQSATQETRALVTQVAQRVEDVWLMVSVKRASLQRAAARPARPVQSVPPQSAALPTPANQQMQIR
Summary
Uniprot
H9IZK7
A0A194PGI0
A0A2H1V143
A0A194QVP0
A0A212EGQ0
A0A1E1WBN5
+ More
A0A1B0GIY9 A0A2J7Q864 A0A1L8DLV9 A0A182MUA3 A0A182PZX4 A0A151IBC3 A0A195BTJ2 F4WV77 A0A151IRI5 A0A195EW97 A0A182IYA6 A0A182X4T8 A0A158NNX2 A0A026WQL9 A0A182I3X8 A0A151WK14 A0A182RB05 U5ESZ7 A0A232FA84 A0A3L8DL03 K7IR14 A0A1A9ZAT3 A0A0C9QUX2 E2C515 A0A2M4CK05 A0A0C9Q0X1 D6W697 A0A2M4BB97 A0A1I8PJL3 A0A182NHA5 A0A182WJ05 T1P8I7 A0A2M4AGM3 A0A182JT36 A0A084VF41 A0A154PP09 W8B2W2 A0A1I8N0J8 A0A0L0C320 A0A2A3E7G9 U4TZ26 A0A1Q3FU42 A0A1Q3FTS9 A0A1Q3FTZ1 A0A310SAF2 A0A087ZVG3 A0A0A1XDK1 A0A0K8W6U2 A0A0K8V9W9 A0A0K8WAG7 A0A1Q3FTS0 A0A0K8VBS4 A0A0Q9XHX7 A0A0K8UGA7 A0A1Y1MLM0 A0A0Q9XM44 A0A0Q9XII7 B4KLR3 A0A0Q9XKF3 A0A1Y1MNF9 A0A1B6JSL1 A0A0R3NKM6 A0A1Y1MPV4 A0A1Y1MJQ8 A0A3B0J773 A0A0Q9WFQ6 A0A2M4BB98 A0A1B0GLU4 A0A182XZA3 A0A0Q9WNQ5 E9J059 A0A182FIB3 A0A0N0BE81 A0A1B6D890 A0A0L7QS96 A0A1J1IKC3 A0A182KS30 A0A067QH20 N6T989 Q5TX53 T1JN60 A0A0N7ZC64 A0A0P5T5Q5 A0A0P5AKL4 A0A0P5DP42 A0A0P6E2T6 A0A0P5SKL0 A0A0P5SV02 A0A0N8CXE0 A0A0P4ZQX2 A0A0P5DTF0
A0A1B0GIY9 A0A2J7Q864 A0A1L8DLV9 A0A182MUA3 A0A182PZX4 A0A151IBC3 A0A195BTJ2 F4WV77 A0A151IRI5 A0A195EW97 A0A182IYA6 A0A182X4T8 A0A158NNX2 A0A026WQL9 A0A182I3X8 A0A151WK14 A0A182RB05 U5ESZ7 A0A232FA84 A0A3L8DL03 K7IR14 A0A1A9ZAT3 A0A0C9QUX2 E2C515 A0A2M4CK05 A0A0C9Q0X1 D6W697 A0A2M4BB97 A0A1I8PJL3 A0A182NHA5 A0A182WJ05 T1P8I7 A0A2M4AGM3 A0A182JT36 A0A084VF41 A0A154PP09 W8B2W2 A0A1I8N0J8 A0A0L0C320 A0A2A3E7G9 U4TZ26 A0A1Q3FU42 A0A1Q3FTS9 A0A1Q3FTZ1 A0A310SAF2 A0A087ZVG3 A0A0A1XDK1 A0A0K8W6U2 A0A0K8V9W9 A0A0K8WAG7 A0A1Q3FTS0 A0A0K8VBS4 A0A0Q9XHX7 A0A0K8UGA7 A0A1Y1MLM0 A0A0Q9XM44 A0A0Q9XII7 B4KLR3 A0A0Q9XKF3 A0A1Y1MNF9 A0A1B6JSL1 A0A0R3NKM6 A0A1Y1MPV4 A0A1Y1MJQ8 A0A3B0J773 A0A0Q9WFQ6 A0A2M4BB98 A0A1B0GLU4 A0A182XZA3 A0A0Q9WNQ5 E9J059 A0A182FIB3 A0A0N0BE81 A0A1B6D890 A0A0L7QS96 A0A1J1IKC3 A0A182KS30 A0A067QH20 N6T989 Q5TX53 T1JN60 A0A0N7ZC64 A0A0P5T5Q5 A0A0P5AKL4 A0A0P5DP42 A0A0P6E2T6 A0A0P5SKL0 A0A0P5SV02 A0A0N8CXE0 A0A0P4ZQX2 A0A0P5DTF0
Pubmed
EMBL
BABH01014374
KQ459606
KPI91814.1
ODYU01000181
SOQ34539.1
KQ461103
+ More
KPJ09389.1 AGBW02015043 OWR40667.1 GDQN01006652 JAT84402.1 AJWK01017910 AJWK01017911 AJWK01017912 NEVH01016978 PNF24765.1 GFDF01006754 JAV07330.1 AXCM01002277 AXCN02000894 KQ978111 KYM96952.1 KQ976417 KYM89751.1 GL888384 EGI61843.1 KQ981129 KYN09271.1 KQ981953 KYN32421.1 ADTU01021865 ADTU01021866 KK107128 EZA58342.1 APCN01000228 KQ983031 KYQ48147.1 GANO01002163 JAB57708.1 NNAY01000622 OXU27343.1 QOIP01000006 RLU21097.1 GBYB01007559 JAG77326.1 GL452714 EFN76969.1 GGFL01001479 MBW65657.1 GBYB01007558 JAG77325.1 KQ971306 EFA11099.2 GGFJ01001183 MBW50324.1 KA644964 AFP59593.1 GGFK01006605 MBW39926.1 ATLV01012360 KE524785 KFB36585.1 KQ435007 KZC13626.1 GAMC01018969 JAB87586.1 JRES01000960 KNC26720.1 KZ288345 PBC27655.1 KB631673 ERL85283.1 GFDL01004049 JAV30996.1 GFDL01004036 JAV31009.1 GFDL01004037 JAV31008.1 KQ762207 OAD56051.1 GBXI01005276 JAD09016.1 GDHF01011579 GDHF01005704 GDHF01001175 JAI40735.1 JAI46610.1 JAI51139.1 GDHF01016979 GDHF01012901 JAI35335.1 JAI39413.1 GDHF01031247 GDHF01018466 GDHF01004494 JAI21067.1 JAI33848.1 JAI47820.1 GFDL01004034 JAV31011.1 GDHF01016324 JAI35990.1 CH933808 KRG04868.1 GDHF01026678 JAI25636.1 GEZM01029713 GEZM01029712 JAV85978.1 KRG04867.1 KRG04866.1 EDW09723.2 KRG04865.1 GEZM01029715 JAV85975.1 GECU01005496 JAT02211.1 CM000071 KRT01435.1 GEZM01029716 GEZM01029714 JAV85976.1 GEZM01029711 JAV85979.1 OUUW01000001 SPP75963.1 CH940648 KRF79867.1 GGFJ01001184 MBW50325.1 AJVK01002046 CH963719 KRF97494.1 GL767291 EFZ13845.1 KQ435845 KOX71191.1 GEDC01015385 JAS21913.1 KQ414766 KOC61409.1 CVRI01000054 CRL00210.1 KK853797 KDQ71697.1 APGK01047262 KB741077 ENN74293.1 AAAB01008807 EAL41713.3 JH431723 GDRN01072309 JAI63576.1 GDIP01132923 JAL70791.1 GDIP01198013 JAJ25389.1 GDIP01159424 JAJ63978.1 GDIQ01068632 JAN26105.1 GDIP01138618 JAL65096.1 GDIQ01099682 JAL52044.1 GDIP01089527 JAM14188.1 GDIP01210060 JAJ13342.1 GDIP01155478 JAJ67924.1
KPJ09389.1 AGBW02015043 OWR40667.1 GDQN01006652 JAT84402.1 AJWK01017910 AJWK01017911 AJWK01017912 NEVH01016978 PNF24765.1 GFDF01006754 JAV07330.1 AXCM01002277 AXCN02000894 KQ978111 KYM96952.1 KQ976417 KYM89751.1 GL888384 EGI61843.1 KQ981129 KYN09271.1 KQ981953 KYN32421.1 ADTU01021865 ADTU01021866 KK107128 EZA58342.1 APCN01000228 KQ983031 KYQ48147.1 GANO01002163 JAB57708.1 NNAY01000622 OXU27343.1 QOIP01000006 RLU21097.1 GBYB01007559 JAG77326.1 GL452714 EFN76969.1 GGFL01001479 MBW65657.1 GBYB01007558 JAG77325.1 KQ971306 EFA11099.2 GGFJ01001183 MBW50324.1 KA644964 AFP59593.1 GGFK01006605 MBW39926.1 ATLV01012360 KE524785 KFB36585.1 KQ435007 KZC13626.1 GAMC01018969 JAB87586.1 JRES01000960 KNC26720.1 KZ288345 PBC27655.1 KB631673 ERL85283.1 GFDL01004049 JAV30996.1 GFDL01004036 JAV31009.1 GFDL01004037 JAV31008.1 KQ762207 OAD56051.1 GBXI01005276 JAD09016.1 GDHF01011579 GDHF01005704 GDHF01001175 JAI40735.1 JAI46610.1 JAI51139.1 GDHF01016979 GDHF01012901 JAI35335.1 JAI39413.1 GDHF01031247 GDHF01018466 GDHF01004494 JAI21067.1 JAI33848.1 JAI47820.1 GFDL01004034 JAV31011.1 GDHF01016324 JAI35990.1 CH933808 KRG04868.1 GDHF01026678 JAI25636.1 GEZM01029713 GEZM01029712 JAV85978.1 KRG04867.1 KRG04866.1 EDW09723.2 KRG04865.1 GEZM01029715 JAV85975.1 GECU01005496 JAT02211.1 CM000071 KRT01435.1 GEZM01029716 GEZM01029714 JAV85976.1 GEZM01029711 JAV85979.1 OUUW01000001 SPP75963.1 CH940648 KRF79867.1 GGFJ01001184 MBW50325.1 AJVK01002046 CH963719 KRF97494.1 GL767291 EFZ13845.1 KQ435845 KOX71191.1 GEDC01015385 JAS21913.1 KQ414766 KOC61409.1 CVRI01000054 CRL00210.1 KK853797 KDQ71697.1 APGK01047262 KB741077 ENN74293.1 AAAB01008807 EAL41713.3 JH431723 GDRN01072309 JAI63576.1 GDIP01132923 JAL70791.1 GDIP01198013 JAJ25389.1 GDIP01159424 JAJ63978.1 GDIQ01068632 JAN26105.1 GDIP01138618 JAL65096.1 GDIQ01099682 JAL52044.1 GDIP01089527 JAM14188.1 GDIP01210060 JAJ13342.1 GDIP01155478 JAJ67924.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000092461
UP000235965
+ More
UP000075883 UP000075886 UP000078542 UP000078540 UP000007755 UP000078492 UP000078541 UP000075880 UP000076407 UP000005205 UP000053097 UP000075840 UP000075809 UP000075900 UP000215335 UP000279307 UP000002358 UP000092445 UP000008237 UP000007266 UP000095300 UP000075884 UP000075920 UP000075881 UP000030765 UP000076502 UP000095301 UP000037069 UP000242457 UP000030742 UP000005203 UP000009192 UP000001819 UP000268350 UP000008792 UP000092462 UP000076408 UP000007798 UP000069272 UP000053105 UP000053825 UP000183832 UP000075882 UP000027135 UP000019118 UP000007062
UP000075883 UP000075886 UP000078542 UP000078540 UP000007755 UP000078492 UP000078541 UP000075880 UP000076407 UP000005205 UP000053097 UP000075840 UP000075809 UP000075900 UP000215335 UP000279307 UP000002358 UP000092445 UP000008237 UP000007266 UP000095300 UP000075884 UP000075920 UP000075881 UP000030765 UP000076502 UP000095301 UP000037069 UP000242457 UP000030742 UP000005203 UP000009192 UP000001819 UP000268350 UP000008792 UP000092462 UP000076408 UP000007798 UP000069272 UP000053105 UP000053825 UP000183832 UP000075882 UP000027135 UP000019118 UP000007062
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IZK7
A0A194PGI0
A0A2H1V143
A0A194QVP0
A0A212EGQ0
A0A1E1WBN5
+ More
A0A1B0GIY9 A0A2J7Q864 A0A1L8DLV9 A0A182MUA3 A0A182PZX4 A0A151IBC3 A0A195BTJ2 F4WV77 A0A151IRI5 A0A195EW97 A0A182IYA6 A0A182X4T8 A0A158NNX2 A0A026WQL9 A0A182I3X8 A0A151WK14 A0A182RB05 U5ESZ7 A0A232FA84 A0A3L8DL03 K7IR14 A0A1A9ZAT3 A0A0C9QUX2 E2C515 A0A2M4CK05 A0A0C9Q0X1 D6W697 A0A2M4BB97 A0A1I8PJL3 A0A182NHA5 A0A182WJ05 T1P8I7 A0A2M4AGM3 A0A182JT36 A0A084VF41 A0A154PP09 W8B2W2 A0A1I8N0J8 A0A0L0C320 A0A2A3E7G9 U4TZ26 A0A1Q3FU42 A0A1Q3FTS9 A0A1Q3FTZ1 A0A310SAF2 A0A087ZVG3 A0A0A1XDK1 A0A0K8W6U2 A0A0K8V9W9 A0A0K8WAG7 A0A1Q3FTS0 A0A0K8VBS4 A0A0Q9XHX7 A0A0K8UGA7 A0A1Y1MLM0 A0A0Q9XM44 A0A0Q9XII7 B4KLR3 A0A0Q9XKF3 A0A1Y1MNF9 A0A1B6JSL1 A0A0R3NKM6 A0A1Y1MPV4 A0A1Y1MJQ8 A0A3B0J773 A0A0Q9WFQ6 A0A2M4BB98 A0A1B0GLU4 A0A182XZA3 A0A0Q9WNQ5 E9J059 A0A182FIB3 A0A0N0BE81 A0A1B6D890 A0A0L7QS96 A0A1J1IKC3 A0A182KS30 A0A067QH20 N6T989 Q5TX53 T1JN60 A0A0N7ZC64 A0A0P5T5Q5 A0A0P5AKL4 A0A0P5DP42 A0A0P6E2T6 A0A0P5SKL0 A0A0P5SV02 A0A0N8CXE0 A0A0P4ZQX2 A0A0P5DTF0
A0A1B0GIY9 A0A2J7Q864 A0A1L8DLV9 A0A182MUA3 A0A182PZX4 A0A151IBC3 A0A195BTJ2 F4WV77 A0A151IRI5 A0A195EW97 A0A182IYA6 A0A182X4T8 A0A158NNX2 A0A026WQL9 A0A182I3X8 A0A151WK14 A0A182RB05 U5ESZ7 A0A232FA84 A0A3L8DL03 K7IR14 A0A1A9ZAT3 A0A0C9QUX2 E2C515 A0A2M4CK05 A0A0C9Q0X1 D6W697 A0A2M4BB97 A0A1I8PJL3 A0A182NHA5 A0A182WJ05 T1P8I7 A0A2M4AGM3 A0A182JT36 A0A084VF41 A0A154PP09 W8B2W2 A0A1I8N0J8 A0A0L0C320 A0A2A3E7G9 U4TZ26 A0A1Q3FU42 A0A1Q3FTS9 A0A1Q3FTZ1 A0A310SAF2 A0A087ZVG3 A0A0A1XDK1 A0A0K8W6U2 A0A0K8V9W9 A0A0K8WAG7 A0A1Q3FTS0 A0A0K8VBS4 A0A0Q9XHX7 A0A0K8UGA7 A0A1Y1MLM0 A0A0Q9XM44 A0A0Q9XII7 B4KLR3 A0A0Q9XKF3 A0A1Y1MNF9 A0A1B6JSL1 A0A0R3NKM6 A0A1Y1MPV4 A0A1Y1MJQ8 A0A3B0J773 A0A0Q9WFQ6 A0A2M4BB98 A0A1B0GLU4 A0A182XZA3 A0A0Q9WNQ5 E9J059 A0A182FIB3 A0A0N0BE81 A0A1B6D890 A0A0L7QS96 A0A1J1IKC3 A0A182KS30 A0A067QH20 N6T989 Q5TX53 T1JN60 A0A0N7ZC64 A0A0P5T5Q5 A0A0P5AKL4 A0A0P5DP42 A0A0P6E2T6 A0A0P5SKL0 A0A0P5SV02 A0A0N8CXE0 A0A0P4ZQX2 A0A0P5DTF0
Ontologies
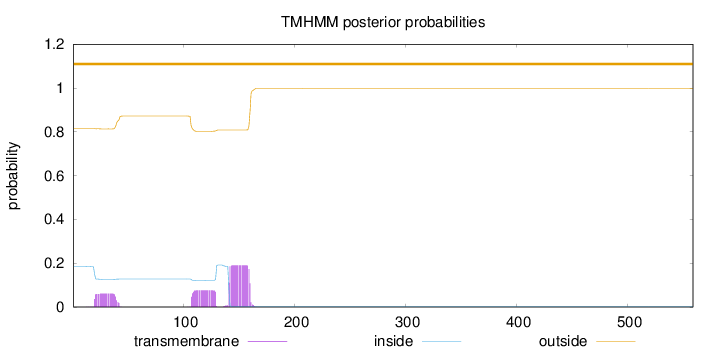
Topology
Length:
559
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.62720999999999
Exp number, first 60 AAs:
1.24314
Total prob of N-in:
0.18447
outside
1 - 559
Population Genetic Test Statistics
Pi
198.72626
Theta
164.379001
Tajima's D
0.483893
CLR
0.550231
CSRT
0.508924553772311
Interpretation
Uncertain
