Gene
KWMTBOMO02530
Pre Gene Modal
BGIBMGA002693
Annotation
PREDICTED:_guanine_nucleotide_exchange_factor_DBS_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.322
Sequence
CDS
ATGTTAAAAACGGGCTATAAAATTGTTAAGGATTTAACTGAGCATCATAAACCTATGGAAACATCTGGCGGTTCGGAGAGTTCCGGCGGTGACGACTCGGATGTTCGACGGTCTCGTCGCGGACACGTTCTAGCTGAACTAATGCAGACCGAGAAAGTATACGTGCAAGAACTCGGATCTATACTCTCTGGGTATAAAGAAGAAATCGAGAAGCCAGAGAACCAGCATCTCCTGCCGCCGACGCTGGTCGGACAGGCGGATGTGTTGTTCGGTAATCTGCACGAACTCTTCACATTCCACCAGGATGTCTTCCTTAAGGACCTGGAGCGATGCATTTCCGCTACAGAACTCGTCGCTTTGTGTTTTGTTGAGAAGAGGGAGACCTTCTTCAGACTGTACAGCTACTACTGTCAGAACATCCCTAGATCGGAGAGGTTGCGGGAGACGTTAGTGGATACGCATCTGTTCCTCCAGGCGTGTCAGCTGCGGCTCGGACACAAGCTGCCGCTGGCTGCGTACCTCCTGAAGCCGGTGCAGAGGATCACTAAATACCAGCTGCTGCTAAAGGATTTACTCCGATACAGCGAGTGTGGATCAATGACGACCGGCCTCCAGCAGGCCTTGGACTGCATGCTCGTCGTATTGAAATGCGTGAATGACTCCATGCACCAGATAGCCATAACCGGAGTCCCTGTGGACTTGAGTCAACAGGGCGAACTCCTGATGCAGGGCTCGTTCCTCGTGGTATCGGAGAACAAGCGCGACCTCCGCCTCAGGATACGGCCCCGGCGACGTCATATCTTCCTCTACGAGAAGGCCATGCTGTTCTGCAAGCCCGCCACAAAGAACAGCCACAATAAAGCCACTTACCACTTCAAACACGATCTGCTGATGTCACAAATAGGTCTTACGGAATCCGTTAAGGGCGATCTCCGCAAGTTCGAGGTGTGGCGCCAGGGCAGGTGTGAGGTGCACACGATCACGGCGCCCACGGTCGAGGTGAAGCGCGCCTGGGTCGAGCGGATCAAACGGGTGCTGCTCGACCAGCTGAAGGAACTCAAGGGCGAGCGAGTTCGGCAGTACGGACTGCAGCATCACAGAACACTAATCCAGACGAGCTCGTGGGAGGTGTCGGGCGCGCAGCCCCGCCCCCAACCCCCCACCGCGCCCCGCACCGGCTCGTGCGACGCGCACGACGAGCCCTGCTGGTCCACGGACCCCTCCGACGACGACGACGACGACGTCGAACACGCACCGAGAACGTGGATATTTCGGTTCTATTCGCTGAACATTAACTATCGTGCGGTGGGATGTGGACTGGTGGCGCTGTCCGACTACACAGCCGTGGGACCGAGCGAAGTGTCGCTGCGAGAGGGACAACACGCTGAACTACTCAAAGTCGGCTGTGCCGGCTGGTGGTACGTGCGGCTTGCAGGCGGACAAGGCGAGGGCTGGGCTCCGGCTGCCTACCTCGACCAGCGCAAGACGTCGCGCTCCTCCAGCAGGTCGCACGACCGCCTTCACGAGCATTGA
Protein
MLKTGYKIVKDLTEHHKPMETSGGSESSGGDDSDVRRSRRGHVLAELMQTEKVYVQELGSILSGYKEEIEKPENQHLLPPTLVGQADVLFGNLHELFTFHQDVFLKDLERCISATELVALCFVEKRETFFRLYSYYCQNIPRSERLRETLVDTHLFLQACQLRLGHKLPLAAYLLKPVQRITKYQLLLKDLLRYSECGSMTTGLQQALDCMLVVLKCVNDSMHQIAITGVPVDLSQQGELLMQGSFLVVSENKRDLRLRIRPRRRHIFLYEKAMLFCKPATKNSHNKATYHFKHDLLMSQIGLTESVKGDLRKFEVWRQGRCEVHTITAPTVEVKRAWVERIKRVLLDQLKELKGERVRQYGLQHHRTLIQTSSWEVSGAQPRPQPPTAPRTGSCDAHDEPCWSTDPSDDDDDDVEHAPRTWIFRFYSLNINYRAVGCGLVALSDYTAVGPSEVSLREGQHAELLKVGCAGWWYVRLAGGQGEGWAPAAYLDQRKTSRSSSRSHDRLHEH
Summary
Uniprot
A0A2A4J0E3
A0A2A4J0I0
A0A194QWQ3
A0A2H1WLV8
A0A212EGQ4
A0A194PEM5
+ More
H9IZK8 A0A2A4J037 A0A0L0C320 A0A1L8DLV9 U5ESZ7 A0A0Q9WFQ6 A0A0Q9W443 A0A0Q9W4H1 A0A0R3NKM6 A0A0Q9W472 A0A0R3NKH8 A0A0R3NKJ1 A0A0R3NKH5 D6W697 A0A2M4CKW2 A0A2M4AJN2 A0A2M3Z8T7 A0A2M3Z8T4 A0A1B0B5C8 A0A084VF41 A0A2M4BB97 A0A0Q9XHX7 A0A0Q9XM44 A0A2M4BB98 A0A0Q9XKF3 A0A3B0J773 Q16L87 A0A2M4AGM3 A0A2M4CK05 A0A0Q9WNQ5 A0A1I8PJP2 A0A1Y1MPV4 A0A182XZA3 A0A1Y1MJQ8 A0A182FIB5 A0A1W4W002 A0A182T4R6 A0A182M338 B4GBJ1 W5JCB5 B3MBR4 E9J059 A0A0L7QS96 A0A154PP09 E2C515 A0A2A3E7G9 A0A310SAF2 A0A026WQL9 A0A3L8DL03 F4WV77 A0A195EW97 A0A195BTJ2 A0A2J7Q864 A0A158NNX2 A0A087ZVG3 A0A151WK14 A0A151IBC3 A0A0C9Q0X1 A0A182RB06 A0A182Q4T4 B4J9F0 E2AEK5 K7IR14 N6T989 A0A0K8UGA7 U4TZ26 A0A034V7T3 A0A034V3F9 A0A182WJ04 E0VRY6 A0A067QH20 A0A182NHA7
H9IZK8 A0A2A4J037 A0A0L0C320 A0A1L8DLV9 U5ESZ7 A0A0Q9WFQ6 A0A0Q9W443 A0A0Q9W4H1 A0A0R3NKM6 A0A0Q9W472 A0A0R3NKH8 A0A0R3NKJ1 A0A0R3NKH5 D6W697 A0A2M4CKW2 A0A2M4AJN2 A0A2M3Z8T7 A0A2M3Z8T4 A0A1B0B5C8 A0A084VF41 A0A2M4BB97 A0A0Q9XHX7 A0A0Q9XM44 A0A2M4BB98 A0A0Q9XKF3 A0A3B0J773 Q16L87 A0A2M4AGM3 A0A2M4CK05 A0A0Q9WNQ5 A0A1I8PJP2 A0A1Y1MPV4 A0A182XZA3 A0A1Y1MJQ8 A0A182FIB5 A0A1W4W002 A0A182T4R6 A0A182M338 B4GBJ1 W5JCB5 B3MBR4 E9J059 A0A0L7QS96 A0A154PP09 E2C515 A0A2A3E7G9 A0A310SAF2 A0A026WQL9 A0A3L8DL03 F4WV77 A0A195EW97 A0A195BTJ2 A0A2J7Q864 A0A158NNX2 A0A087ZVG3 A0A151WK14 A0A151IBC3 A0A0C9Q0X1 A0A182RB06 A0A182Q4T4 B4J9F0 E2AEK5 K7IR14 N6T989 A0A0K8UGA7 U4TZ26 A0A034V7T3 A0A034V3F9 A0A182WJ04 E0VRY6 A0A067QH20 A0A182NHA7
Pubmed
EMBL
NWSH01004591
PCG64882.1
PCG64883.1
KQ461103
KPJ09390.1
ODYU01009553
+ More
SOQ54061.1 AGBW02015043 OWR40669.1 KQ459606 KPI91811.1 BABH01014380 BABH01014381 BABH01014382 PCG64884.1 JRES01000960 KNC26720.1 GFDF01006754 JAV07330.1 GANO01002163 JAB57708.1 CH940648 KRF79867.1 KRF79865.1 KRF79864.1 CM000071 KRT01435.1 KRF79863.1 KRF79866.1 KRT01433.1 KRT01431.1 KRT01434.1 KQ971306 EFA11099.2 GGFL01001798 MBW65976.1 GGFK01007631 MBW40952.1 GGFM01004195 MBW24946.1 GGFM01004196 MBW24947.1 JXJN01008672 ATLV01012360 KE524785 KFB36585.1 GGFJ01001183 MBW50324.1 CH933808 KRG04868.1 KRG04867.1 GGFJ01001184 MBW50325.1 KRG04865.1 OUUW01000001 SPP75963.1 CH477917 EAT35078.1 GGFK01006605 MBW39926.1 GGFL01001479 MBW65657.1 CH963719 KRF97494.1 GEZM01029716 GEZM01029714 JAV85976.1 GEZM01029711 JAV85979.1 AXCM01015712 CH479181 EDW31286.1 ADMH02001929 ETN60469.1 CH902619 EDV38210.2 GL767291 EFZ13845.1 KQ414766 KOC61409.1 KQ435007 KZC13626.1 GL452714 EFN76969.1 KZ288345 PBC27655.1 KQ762207 OAD56051.1 KK107128 EZA58342.1 QOIP01000006 RLU21097.1 GL888384 EGI61843.1 KQ981953 KYN32421.1 KQ976417 KYM89751.1 NEVH01016978 PNF24765.1 ADTU01021865 ADTU01021866 KQ983031 KYQ48147.1 KQ978111 KYM96952.1 GBYB01007558 JAG77325.1 AXCN02000894 CH916367 EDW01431.1 GL438870 EFN68128.1 APGK01047262 KB741077 ENN74293.1 GDHF01026678 JAI25636.1 KB631673 ERL85283.1 GAKP01021137 JAC37815.1 GAKP01021136 JAC37816.1 DS235634 EEB16142.1 KK853797 KDQ71697.1
SOQ54061.1 AGBW02015043 OWR40669.1 KQ459606 KPI91811.1 BABH01014380 BABH01014381 BABH01014382 PCG64884.1 JRES01000960 KNC26720.1 GFDF01006754 JAV07330.1 GANO01002163 JAB57708.1 CH940648 KRF79867.1 KRF79865.1 KRF79864.1 CM000071 KRT01435.1 KRF79863.1 KRF79866.1 KRT01433.1 KRT01431.1 KRT01434.1 KQ971306 EFA11099.2 GGFL01001798 MBW65976.1 GGFK01007631 MBW40952.1 GGFM01004195 MBW24946.1 GGFM01004196 MBW24947.1 JXJN01008672 ATLV01012360 KE524785 KFB36585.1 GGFJ01001183 MBW50324.1 CH933808 KRG04868.1 KRG04867.1 GGFJ01001184 MBW50325.1 KRG04865.1 OUUW01000001 SPP75963.1 CH477917 EAT35078.1 GGFK01006605 MBW39926.1 GGFL01001479 MBW65657.1 CH963719 KRF97494.1 GEZM01029716 GEZM01029714 JAV85976.1 GEZM01029711 JAV85979.1 AXCM01015712 CH479181 EDW31286.1 ADMH02001929 ETN60469.1 CH902619 EDV38210.2 GL767291 EFZ13845.1 KQ414766 KOC61409.1 KQ435007 KZC13626.1 GL452714 EFN76969.1 KZ288345 PBC27655.1 KQ762207 OAD56051.1 KK107128 EZA58342.1 QOIP01000006 RLU21097.1 GL888384 EGI61843.1 KQ981953 KYN32421.1 KQ976417 KYM89751.1 NEVH01016978 PNF24765.1 ADTU01021865 ADTU01021866 KQ983031 KYQ48147.1 KQ978111 KYM96952.1 GBYB01007558 JAG77325.1 AXCN02000894 CH916367 EDW01431.1 GL438870 EFN68128.1 APGK01047262 KB741077 ENN74293.1 GDHF01026678 JAI25636.1 KB631673 ERL85283.1 GAKP01021137 JAC37815.1 GAKP01021136 JAC37816.1 DS235634 EEB16142.1 KK853797 KDQ71697.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000005204
UP000037069
+ More
UP000008792 UP000001819 UP000007266 UP000092460 UP000030765 UP000009192 UP000268350 UP000008820 UP000007798 UP000095300 UP000076408 UP000069272 UP000192221 UP000075901 UP000075883 UP000008744 UP000000673 UP000007801 UP000053825 UP000076502 UP000008237 UP000242457 UP000053097 UP000279307 UP000007755 UP000078541 UP000078540 UP000235965 UP000005205 UP000005203 UP000075809 UP000078542 UP000075900 UP000075886 UP000001070 UP000000311 UP000002358 UP000019118 UP000030742 UP000075920 UP000009046 UP000027135 UP000075884
UP000008792 UP000001819 UP000007266 UP000092460 UP000030765 UP000009192 UP000268350 UP000008820 UP000007798 UP000095300 UP000076408 UP000069272 UP000192221 UP000075901 UP000075883 UP000008744 UP000000673 UP000007801 UP000053825 UP000076502 UP000008237 UP000242457 UP000053097 UP000279307 UP000007755 UP000078541 UP000078540 UP000235965 UP000005205 UP000005203 UP000075809 UP000078542 UP000075900 UP000075886 UP000001070 UP000000311 UP000002358 UP000019118 UP000030742 UP000075920 UP000009046 UP000027135 UP000075884
PRIDE
Interpro
IPR001331
GDS_CDC24_CS
+ More
IPR011993 PH-like_dom_sf
IPR000219 DH-domain
IPR001849 PH_domain
IPR036028 SH3-like_dom_sf
IPR001452 SH3_domain
IPR035899 DBL_dom_sf
IPR001251 CRAL-TRIO_dom
IPR036865 CRAL-TRIO_dom_sf
IPR018159 Spectrin/alpha-actinin
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR011993 PH-like_dom_sf
IPR000219 DH-domain
IPR001849 PH_domain
IPR036028 SH3-like_dom_sf
IPR001452 SH3_domain
IPR035899 DBL_dom_sf
IPR001251 CRAL-TRIO_dom
IPR036865 CRAL-TRIO_dom_sf
IPR018159 Spectrin/alpha-actinin
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
Gene 3D
ProteinModelPortal
A0A2A4J0E3
A0A2A4J0I0
A0A194QWQ3
A0A2H1WLV8
A0A212EGQ4
A0A194PEM5
+ More
H9IZK8 A0A2A4J037 A0A0L0C320 A0A1L8DLV9 U5ESZ7 A0A0Q9WFQ6 A0A0Q9W443 A0A0Q9W4H1 A0A0R3NKM6 A0A0Q9W472 A0A0R3NKH8 A0A0R3NKJ1 A0A0R3NKH5 D6W697 A0A2M4CKW2 A0A2M4AJN2 A0A2M3Z8T7 A0A2M3Z8T4 A0A1B0B5C8 A0A084VF41 A0A2M4BB97 A0A0Q9XHX7 A0A0Q9XM44 A0A2M4BB98 A0A0Q9XKF3 A0A3B0J773 Q16L87 A0A2M4AGM3 A0A2M4CK05 A0A0Q9WNQ5 A0A1I8PJP2 A0A1Y1MPV4 A0A182XZA3 A0A1Y1MJQ8 A0A182FIB5 A0A1W4W002 A0A182T4R6 A0A182M338 B4GBJ1 W5JCB5 B3MBR4 E9J059 A0A0L7QS96 A0A154PP09 E2C515 A0A2A3E7G9 A0A310SAF2 A0A026WQL9 A0A3L8DL03 F4WV77 A0A195EW97 A0A195BTJ2 A0A2J7Q864 A0A158NNX2 A0A087ZVG3 A0A151WK14 A0A151IBC3 A0A0C9Q0X1 A0A182RB06 A0A182Q4T4 B4J9F0 E2AEK5 K7IR14 N6T989 A0A0K8UGA7 U4TZ26 A0A034V7T3 A0A034V3F9 A0A182WJ04 E0VRY6 A0A067QH20 A0A182NHA7
H9IZK8 A0A2A4J037 A0A0L0C320 A0A1L8DLV9 U5ESZ7 A0A0Q9WFQ6 A0A0Q9W443 A0A0Q9W4H1 A0A0R3NKM6 A0A0Q9W472 A0A0R3NKH8 A0A0R3NKJ1 A0A0R3NKH5 D6W697 A0A2M4CKW2 A0A2M4AJN2 A0A2M3Z8T7 A0A2M3Z8T4 A0A1B0B5C8 A0A084VF41 A0A2M4BB97 A0A0Q9XHX7 A0A0Q9XM44 A0A2M4BB98 A0A0Q9XKF3 A0A3B0J773 Q16L87 A0A2M4AGM3 A0A2M4CK05 A0A0Q9WNQ5 A0A1I8PJP2 A0A1Y1MPV4 A0A182XZA3 A0A1Y1MJQ8 A0A182FIB5 A0A1W4W002 A0A182T4R6 A0A182M338 B4GBJ1 W5JCB5 B3MBR4 E9J059 A0A0L7QS96 A0A154PP09 E2C515 A0A2A3E7G9 A0A310SAF2 A0A026WQL9 A0A3L8DL03 F4WV77 A0A195EW97 A0A195BTJ2 A0A2J7Q864 A0A158NNX2 A0A087ZVG3 A0A151WK14 A0A151IBC3 A0A0C9Q0X1 A0A182RB06 A0A182Q4T4 B4J9F0 E2AEK5 K7IR14 N6T989 A0A0K8UGA7 U4TZ26 A0A034V7T3 A0A034V3F9 A0A182WJ04 E0VRY6 A0A067QH20 A0A182NHA7
PDB
1KZ7
E-value=3.4025e-75,
Score=718
Ontologies
GO
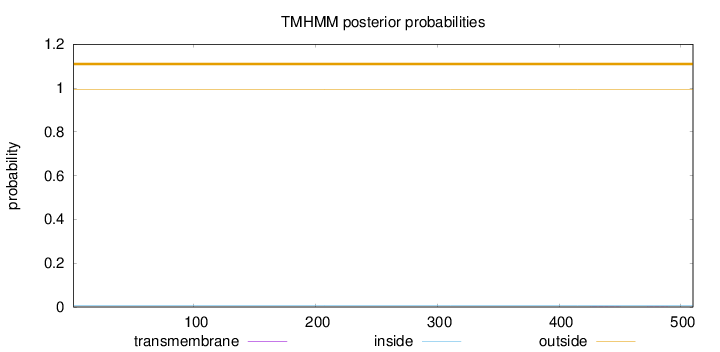
Topology
Length:
510
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01407
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00553
outside
1 - 510
Population Genetic Test Statistics
Pi
272.527369
Theta
189.266705
Tajima's D
1.453977
CLR
0.398735
CSRT
0.777661116944153
Interpretation
Uncertain
