Gene
KWMTBOMO02529 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002594
Annotation
adenylate_kinase_2_[Bombyx_mori]
Full name
Adenylate kinase
Alternative Name
ATP-AMP transphosphorylase
ATP:AMP phosphotransferase
Adenylate kinase cytosolic and mitochondrial
Adenylate monophosphate kinase
ATP:AMP phosphotransferase
Adenylate kinase cytosolic and mitochondrial
Adenylate monophosphate kinase
Location in the cell
Cytoplasmic Reliability : 1.345 Mitochondrial Reliability : 1.775 Nuclear Reliability : 1.154
Sequence
CDS
ATGGCACCGGCCGCTGCAGCCACGAAACTAAAACCTGATGAGGATCCTTTAGGAATAAGGGCCGTGCTACTTGGCCCTCCTGGATCCGGTAAGGGGACTCAGGCTCCTCGTCTGAAAGAAAAGTATTGTGTTTGTCACCTATCCACTGGTGACATGCTGCGTGCTGAAGTCTCATCGGGATCAGACCTGGGACGACGCCTTAAGAAGGTCATGGATGAGGGCAAACTTGTTTCCGATGAAATGGTGGTTGATATGATTGACAAGAACTTGGATCAACCTGAGTGCAAGAATGGTTTCTTACTTGATGGATTCCCTAGAACTGTACCCCAGGCTGAGAAGTTGGACGATCTATTGGCGAAGCGCAAGACTGCACTGGACGCCGTAATCGAGTTCGGAATCGAGGACAGTCTGCTCGTAAGGAGAATTACGGGCAGACTGATTCACCCACCTAGTGGTCGTAGCTATCACGAAGAGTTCCACCCGCCGAAGAAACCAATGACTGACGATGTCACCGGAGAAGCTTTGATAAAAAGGTCCGACGACAATGTCGAGGCTCTGAAGAAACGCTTAGCTACATATCATGCTCAAACGGTTCCTTTAGTAGACTATTACATGAGGAAAGGTCTACATTGGCGCGTCGATGCATCGAAAGCCGCTGACGACGTCTTCAACAAAATCGATAATATCTTTAAGAGTAGAATTTTCAGCAGAGCACGGTCTGCTTTGTAA
Protein
MAPAAAATKLKPDEDPLGIRAVLLGPPGSGKGTQAPRLKEKYCVCHLSTGDMLRAEVSSGSDLGRRLKKVMDEGKLVSDEMVVDMIDKNLDQPECKNGFLLDGFPRTVPQAEKLDDLLAKRKTALDAVIEFGIEDSLLVRRITGRLIHPPSGRSYHEEFHPPKKPMTDDVTGEALIKRSDDNVEALKKRLATYHAQTVPLVDYYMRKGLHWRVDASKAADDVFNKIDNIFKSRIFSRARSAL
Summary
Description
Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. Plays an important role in cellular energy homeostasis and in adenine nucleotide metabolism. Adenylate kinase activity is critical for regulation of the phosphate utilization and the AMP de novo biosynthesis pathways.
Catalytic Activity
AMP + ATP = 2 ADP
Subunit
Monomer.
Similarity
Belongs to the adenylate kinase family. AK2 subfamily.
Belongs to the adenylate kinase family.
Belongs to the adenylate kinase family.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Kinase
Mitochondrion
Nucleotide-binding
Reference proteome
Transferase
Phosphoprotein
Feature
chain Adenylate kinase
Uniprot
D2Y062
I7E453
A0A194R0M3
A0A194PL28
I1XB05
S4PGE2
+ More
A0A2A4IYP6 A0A0L7LIT0 A0A2H1WLV9 A0A212EGS0 D6W7C6 J3JTL3 A0A1B6F5R1 W8CEH7 N6U870 A0A182UUR3 A0A182JT33 A0A182TDG5 A0A1B0C953 A0A182X4T6 A0A182KS19 A0A182I3X5 Q7QJX9 A0A1A9ZZH7 A0A336M5Z7 A0A1B6HC04 A0A1B6M664 A0A182P4G3 A0A182IYG8 A0A2M3ZI41 A0A1A9V699 W5JSV0 B4PAR6 D3TLM5 A0A1L8EEJ1 A0A0J9RL87 B4QBH8 B4I2A8 B3NQ53 A0A034VQD5 A0A2M4AMT8 A0A0L0BQJ4 Q9U915 T1PER8 B4MQT3 A0A182MTV2 A0A182GVU5 A0A1B0BW27 A0A023EKV1 Q1HQK0 A0A1I8PDA2 A0A1W4UZE1 A0A0A1WU31 A0A1Q3EXB4 A0A182RB07 V5G5Q0 A0A084VF42 A0A1L8DXZ1 A0A2M4BY64 B0X5E3 A0A1A9WI61 A0A1Y1LBS1 A0A182NHA8 B3MCQ5 A0A182QXH5 A0A1L8DXV0 A0A182WJ02 A0A182XZA4 A0A0K8TT75 T1E828 A0A087SUP8 B4KLY1 A0A1B6CWT8 B4LP08 A0A151IBS1 B4J672 U5EXT6 A0A0L7QRZ4 A0A0P4X9D0 A0A0P5J3E6 A0A0P5J980 A0A0P6IBS3 Q290A8 A0A0P4WVG2 A0A2S2NUU7 A0A0M4EBJ7 E9GFK4 F4WV76 A0A0B4J2Q4 E9J058 A0A151WJW2 A0A0C9R051 A0A3B0JDA8 A0A026WRM3 A0A195EVX7 A0A2H8TX84 A0A1W4X547 A0A151IRE1 C4WT06 A0A1J1IRY8
A0A2A4IYP6 A0A0L7LIT0 A0A2H1WLV9 A0A212EGS0 D6W7C6 J3JTL3 A0A1B6F5R1 W8CEH7 N6U870 A0A182UUR3 A0A182JT33 A0A182TDG5 A0A1B0C953 A0A182X4T6 A0A182KS19 A0A182I3X5 Q7QJX9 A0A1A9ZZH7 A0A336M5Z7 A0A1B6HC04 A0A1B6M664 A0A182P4G3 A0A182IYG8 A0A2M3ZI41 A0A1A9V699 W5JSV0 B4PAR6 D3TLM5 A0A1L8EEJ1 A0A0J9RL87 B4QBH8 B4I2A8 B3NQ53 A0A034VQD5 A0A2M4AMT8 A0A0L0BQJ4 Q9U915 T1PER8 B4MQT3 A0A182MTV2 A0A182GVU5 A0A1B0BW27 A0A023EKV1 Q1HQK0 A0A1I8PDA2 A0A1W4UZE1 A0A0A1WU31 A0A1Q3EXB4 A0A182RB07 V5G5Q0 A0A084VF42 A0A1L8DXZ1 A0A2M4BY64 B0X5E3 A0A1A9WI61 A0A1Y1LBS1 A0A182NHA8 B3MCQ5 A0A182QXH5 A0A1L8DXV0 A0A182WJ02 A0A182XZA4 A0A0K8TT75 T1E828 A0A087SUP8 B4KLY1 A0A1B6CWT8 B4LP08 A0A151IBS1 B4J672 U5EXT6 A0A0L7QRZ4 A0A0P4X9D0 A0A0P5J3E6 A0A0P5J980 A0A0P6IBS3 Q290A8 A0A0P4WVG2 A0A2S2NUU7 A0A0M4EBJ7 E9GFK4 F4WV76 A0A0B4J2Q4 E9J058 A0A151WJW2 A0A0C9R051 A0A3B0JDA8 A0A026WRM3 A0A195EVX7 A0A2H8TX84 A0A1W4X547 A0A151IRE1 C4WT06 A0A1J1IRY8
EC Number
2.7.4.3
Pubmed
19121390
26354079
23622113
26227816
22118469
18362917
+ More
19820115 22516182 24495485 23537049 20966253 12364791 20920257 23761445 17994087 20353571 22936249 25348373 26108605 10786623 10731132 12537572 18327897 25315136 26483478 24945155 17204158 17510324 25830018 24438588 28004739 25244985 26369729 15632085 21292972 21719571 20075255 21282665 24508170 30249741
19820115 22516182 24495485 23537049 20966253 12364791 20920257 23761445 17994087 20353571 22936249 25348373 26108605 10786623 10731132 12537572 18327897 25315136 26483478 24945155 17204158 17510324 25830018 24438588 28004739 25244985 26369729 15632085 21292972 21719571 20075255 21282665 24508170 30249741
EMBL
BABH01014382
BABH01014383
GU270853
ADB27115.1
JX046921
AFO83997.1
+ More
KQ461103 KPJ09391.1 KQ459606 KPI91810.1 JQ801344 AFI80907.1 GAIX01003707 JAA88853.1 NWSH01004591 PCG64881.1 JTDY01000942 KOB75352.1 ODYU01009553 SOQ54060.1 AGBW02015043 OWR40670.1 KQ971307 EFA11124.1 BT126570 AEE61534.1 GECZ01024252 JAS45517.1 GAMC01000176 JAC06380.1 APGK01039127 KB740967 KB632399 ENN76846.1 ERL94665.1 AJWK01001915 AJWK01001916 APCN01000227 AAAB01008807 UFQT01000381 SSX23787.1 GECU01035489 GECU01025473 GECU01015473 JAS72217.1 JAS82233.1 JAS92233.1 GEBQ01008584 JAT31393.1 GGFM01007431 MBW28182.1 ADMH02000542 ETN65995.1 CM000158 CCAG010018993 EZ422327 ADD18603.1 GFDG01001662 JAV17137.1 CM002911 KMY96219.1 CM000362 CH480820 CH480824 CH954179 GAKP01013431 JAC45521.1 GGFK01008768 MBW42089.1 JRES01001518 KNC22281.1 AB009996 AE013599 AY069848 KA647277 AFP61906.1 CH963849 AXCM01015711 JXUM01020521 KQ560574 KXJ81847.1 JXJN01021586 GAPW01003801 JAC09797.1 DQ440444 CH477917 GBXI01012369 JAD01923.1 GFDL01015088 JAV19957.1 GALX01003061 JAB65405.1 ATLV01012360 KE524785 KFB36586.1 GFDF01002822 JAV11262.1 GGFJ01008884 MBW58025.1 DS232375 GEZM01060343 JAV71053.1 CH902619 AXCN02000894 GFDF01002850 JAV11234.1 GDAI01000252 JAI17351.1 GAMD01002562 JAA99028.1 KK112039 KFM56587.1 CH933808 GEDC01019329 JAS17969.1 CH940648 KQ978111 KYM96948.1 CH916367 GANO01000763 JAB59108.1 KQ414766 KOC61410.1 GDIP01245812 JAI77589.1 GDIQ01205224 JAK46501.1 GDIQ01203476 JAK48249.1 GDIQ01023231 JAN71506.1 CM000071 GDIP01254596 JAI68805.1 GGMR01008193 MBY20812.1 CP012524 ALC42646.1 GL732542 EFX81793.1 GL888384 EGI61842.1 GL767291 EFZ13829.1 KQ983031 KYQ48148.1 GBYB01000216 JAG69983.1 OUUW01000001 SPP73210.1 KK107128 QOIP01000006 EZA58341.1 RLU21104.1 KQ981953 KYN32420.1 GFXV01007062 MBW18867.1 KQ981129 KYN09272.1 ABLF02021004 AK340409 BAH71026.1 CVRI01000059 CRL02983.1
KQ461103 KPJ09391.1 KQ459606 KPI91810.1 JQ801344 AFI80907.1 GAIX01003707 JAA88853.1 NWSH01004591 PCG64881.1 JTDY01000942 KOB75352.1 ODYU01009553 SOQ54060.1 AGBW02015043 OWR40670.1 KQ971307 EFA11124.1 BT126570 AEE61534.1 GECZ01024252 JAS45517.1 GAMC01000176 JAC06380.1 APGK01039127 KB740967 KB632399 ENN76846.1 ERL94665.1 AJWK01001915 AJWK01001916 APCN01000227 AAAB01008807 UFQT01000381 SSX23787.1 GECU01035489 GECU01025473 GECU01015473 JAS72217.1 JAS82233.1 JAS92233.1 GEBQ01008584 JAT31393.1 GGFM01007431 MBW28182.1 ADMH02000542 ETN65995.1 CM000158 CCAG010018993 EZ422327 ADD18603.1 GFDG01001662 JAV17137.1 CM002911 KMY96219.1 CM000362 CH480820 CH480824 CH954179 GAKP01013431 JAC45521.1 GGFK01008768 MBW42089.1 JRES01001518 KNC22281.1 AB009996 AE013599 AY069848 KA647277 AFP61906.1 CH963849 AXCM01015711 JXUM01020521 KQ560574 KXJ81847.1 JXJN01021586 GAPW01003801 JAC09797.1 DQ440444 CH477917 GBXI01012369 JAD01923.1 GFDL01015088 JAV19957.1 GALX01003061 JAB65405.1 ATLV01012360 KE524785 KFB36586.1 GFDF01002822 JAV11262.1 GGFJ01008884 MBW58025.1 DS232375 GEZM01060343 JAV71053.1 CH902619 AXCN02000894 GFDF01002850 JAV11234.1 GDAI01000252 JAI17351.1 GAMD01002562 JAA99028.1 KK112039 KFM56587.1 CH933808 GEDC01019329 JAS17969.1 CH940648 KQ978111 KYM96948.1 CH916367 GANO01000763 JAB59108.1 KQ414766 KOC61410.1 GDIP01245812 JAI77589.1 GDIQ01205224 JAK46501.1 GDIQ01203476 JAK48249.1 GDIQ01023231 JAN71506.1 CM000071 GDIP01254596 JAI68805.1 GGMR01008193 MBY20812.1 CP012524 ALC42646.1 GL732542 EFX81793.1 GL888384 EGI61842.1 GL767291 EFZ13829.1 KQ983031 KYQ48148.1 GBYB01000216 JAG69983.1 OUUW01000001 SPP73210.1 KK107128 QOIP01000006 EZA58341.1 RLU21104.1 KQ981953 KYN32420.1 GFXV01007062 MBW18867.1 KQ981129 KYN09272.1 ABLF02021004 AK340409 BAH71026.1 CVRI01000059 CRL02983.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000007266 UP000019118 UP000030742 UP000075903 UP000075881 UP000075902 UP000092461 UP000076407 UP000075882 UP000075840 UP000007062 UP000092445 UP000075885 UP000075880 UP000078200 UP000000673 UP000002282 UP000092444 UP000000304 UP000001292 UP000008711 UP000037069 UP000000803 UP000095301 UP000007798 UP000075883 UP000069940 UP000249989 UP000092460 UP000008820 UP000095300 UP000192221 UP000075900 UP000030765 UP000002320 UP000091820 UP000075884 UP000007801 UP000075886 UP000075920 UP000076408 UP000054359 UP000009192 UP000008792 UP000078542 UP000001070 UP000053825 UP000001819 UP000092553 UP000000305 UP000007755 UP000002358 UP000075809 UP000268350 UP000053097 UP000279307 UP000078541 UP000192223 UP000078492 UP000007819 UP000183832
UP000007266 UP000019118 UP000030742 UP000075903 UP000075881 UP000075902 UP000092461 UP000076407 UP000075882 UP000075840 UP000007062 UP000092445 UP000075885 UP000075880 UP000078200 UP000000673 UP000002282 UP000092444 UP000000304 UP000001292 UP000008711 UP000037069 UP000000803 UP000095301 UP000007798 UP000075883 UP000069940 UP000249989 UP000092460 UP000008820 UP000095300 UP000192221 UP000075900 UP000030765 UP000002320 UP000091820 UP000075884 UP000007801 UP000075886 UP000075920 UP000076408 UP000054359 UP000009192 UP000008792 UP000078542 UP000001070 UP000053825 UP000001819 UP000092553 UP000000305 UP000007755 UP000002358 UP000075809 UP000268350 UP000053097 UP000279307 UP000078541 UP000192223 UP000078492 UP000007819 UP000183832
Pfam
PF05191 ADK_lid
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
D2Y062
I7E453
A0A194R0M3
A0A194PL28
I1XB05
S4PGE2
+ More
A0A2A4IYP6 A0A0L7LIT0 A0A2H1WLV9 A0A212EGS0 D6W7C6 J3JTL3 A0A1B6F5R1 W8CEH7 N6U870 A0A182UUR3 A0A182JT33 A0A182TDG5 A0A1B0C953 A0A182X4T6 A0A182KS19 A0A182I3X5 Q7QJX9 A0A1A9ZZH7 A0A336M5Z7 A0A1B6HC04 A0A1B6M664 A0A182P4G3 A0A182IYG8 A0A2M3ZI41 A0A1A9V699 W5JSV0 B4PAR6 D3TLM5 A0A1L8EEJ1 A0A0J9RL87 B4QBH8 B4I2A8 B3NQ53 A0A034VQD5 A0A2M4AMT8 A0A0L0BQJ4 Q9U915 T1PER8 B4MQT3 A0A182MTV2 A0A182GVU5 A0A1B0BW27 A0A023EKV1 Q1HQK0 A0A1I8PDA2 A0A1W4UZE1 A0A0A1WU31 A0A1Q3EXB4 A0A182RB07 V5G5Q0 A0A084VF42 A0A1L8DXZ1 A0A2M4BY64 B0X5E3 A0A1A9WI61 A0A1Y1LBS1 A0A182NHA8 B3MCQ5 A0A182QXH5 A0A1L8DXV0 A0A182WJ02 A0A182XZA4 A0A0K8TT75 T1E828 A0A087SUP8 B4KLY1 A0A1B6CWT8 B4LP08 A0A151IBS1 B4J672 U5EXT6 A0A0L7QRZ4 A0A0P4X9D0 A0A0P5J3E6 A0A0P5J980 A0A0P6IBS3 Q290A8 A0A0P4WVG2 A0A2S2NUU7 A0A0M4EBJ7 E9GFK4 F4WV76 A0A0B4J2Q4 E9J058 A0A151WJW2 A0A0C9R051 A0A3B0JDA8 A0A026WRM3 A0A195EVX7 A0A2H8TX84 A0A1W4X547 A0A151IRE1 C4WT06 A0A1J1IRY8
A0A2A4IYP6 A0A0L7LIT0 A0A2H1WLV9 A0A212EGS0 D6W7C6 J3JTL3 A0A1B6F5R1 W8CEH7 N6U870 A0A182UUR3 A0A182JT33 A0A182TDG5 A0A1B0C953 A0A182X4T6 A0A182KS19 A0A182I3X5 Q7QJX9 A0A1A9ZZH7 A0A336M5Z7 A0A1B6HC04 A0A1B6M664 A0A182P4G3 A0A182IYG8 A0A2M3ZI41 A0A1A9V699 W5JSV0 B4PAR6 D3TLM5 A0A1L8EEJ1 A0A0J9RL87 B4QBH8 B4I2A8 B3NQ53 A0A034VQD5 A0A2M4AMT8 A0A0L0BQJ4 Q9U915 T1PER8 B4MQT3 A0A182MTV2 A0A182GVU5 A0A1B0BW27 A0A023EKV1 Q1HQK0 A0A1I8PDA2 A0A1W4UZE1 A0A0A1WU31 A0A1Q3EXB4 A0A182RB07 V5G5Q0 A0A084VF42 A0A1L8DXZ1 A0A2M4BY64 B0X5E3 A0A1A9WI61 A0A1Y1LBS1 A0A182NHA8 B3MCQ5 A0A182QXH5 A0A1L8DXV0 A0A182WJ02 A0A182XZA4 A0A0K8TT75 T1E828 A0A087SUP8 B4KLY1 A0A1B6CWT8 B4LP08 A0A151IBS1 B4J672 U5EXT6 A0A0L7QRZ4 A0A0P4X9D0 A0A0P5J3E6 A0A0P5J980 A0A0P6IBS3 Q290A8 A0A0P4WVG2 A0A2S2NUU7 A0A0M4EBJ7 E9GFK4 F4WV76 A0A0B4J2Q4 E9J058 A0A151WJW2 A0A0C9R051 A0A3B0JDA8 A0A026WRM3 A0A195EVX7 A0A2H8TX84 A0A1W4X547 A0A151IRE1 C4WT06 A0A1J1IRY8
PDB
2C9Y
E-value=6.56918e-86,
Score=806
Ontologies
PATHWAY
GO
PANTHER
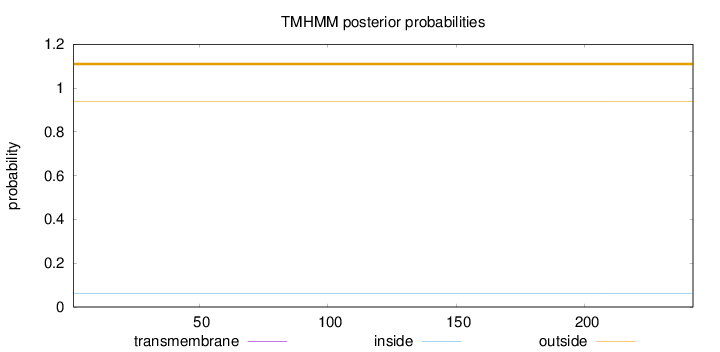
Topology
Subcellular location
Cytoplasm
Predominantly mitochondrial. With evidence from 1 publications.
Cytosol Predominantly mitochondrial. With evidence from 1 publications.
Mitochondrion intermembrane space Predominantly mitochondrial. With evidence from 1 publications.
Cytosol Predominantly mitochondrial. With evidence from 1 publications.
Mitochondrion intermembrane space Predominantly mitochondrial. With evidence from 1 publications.
Length:
242
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06062
outside
1 - 242
Population Genetic Test Statistics
Pi
175.405468
Theta
193.444821
Tajima's D
-0.389902
CLR
0.340935
CSRT
0.260836958152092
Interpretation
Uncertain
