Gene
KWMTBOMO02525 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002593
Annotation
PREDICTED:_prolyl_endopeptidase_isoform_X1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.272
Sequence
CDS
ATGGGCTTGGTGTTCGCGATGCGTGCCTGTATAATATGTGTTTTAATAGCGTGCGCCTTTTTGCAGTGCACCGGCACGACAATATTGTCCTCAAGGAATATGCTGTATGACTATCCAGCGGTGAGGCGTGACGAAACCGTCGTCGACGACTATCATGGAACTAAGATCAAAGATCCATACCGTTGGCTCGAAGATCCAGATTCGAATGAAACCAAAGAATTCGTCGAGGCTCAGAACAAGATAACGCGTCCGTATTTAGACGCGTGTCCCGTGCAGAAATCGATCAACGAGAGGCTGACGGAACTGTGGAACTACCCTAAATATTCATGTCCCTTCAGGAAGGGCAGCAGATACTTCTTCTTCAAGAACACCGGTCTGCAGAACCAAAATGCGTTGTACGTGCAAGATGGTCTCGACGGCGAACCTCGCGTCTTCTTGGACCCGAACACTCTGTCCGAAGACGGCACGGTGGCGCTTTCCGGTACCAAGTTCACCGAAGACGGATCCACCTTGGCGTACGGACTTTCTGCCAGCGGCTCTGATTGGATAGCTATACATTTCAAAGACGTTGCCACCGGCGAGGATTATCCGGAGGTTTTAGAAAAAGTCAAATTCGCTTCGATGTCGTGGACGAAAGATAACAAGGGATTGTTTTACTCGATGTACCCGAAGCAGACCGGCAAGACGGACGGATCGGAAACTGAAGTGAATCACGACCAGAAACTCTGCTATCACCGGATAAACACGTCGCAGTCCGAAGACGTTGTCGTTGTGGAGTTTCCTGAGGAACCGCTGTGGAGAATCGTAGCCGACGTGTCTGATTGCGGCCGGTACTTGCTCGTGTACCCGGTGCGCGACAGCCGCGACAACCTGCTGTTCTTCGCCGACCTCAGCAAGCACCCGGAAATCAACGGGAAGCTGCCGCTCATGCCAGTCGTCGAGGAATTCGAAGCCGATTACGAGTACGTCACGAATGAGGATTCTGTATGCATCATCCGCACGAACAAGAATGCTCCCAACTACAGGCTGATCATGATCGACCTCCAGAATCCCGCCAAGGAAAACTGGCGGACGTTAGTACCCGAACACCCGACGGACGTGCTGGATTGGGCGGCCGCCGTAGACCAAGATAAACTGGTCATACACTACATCAGGGACGTAAAGAGCGTACTTCAATTGCACGACTTGAAAACTGGCGAATTTATACAAACGTTTCCGTTGGACGTCGGCTCCGTGGTCGGCTTCACCGGCAAGAAGAATCAAACGGAAATATTCTATCACTTCATGTCGTTCCTCACGCCGGGAGTCATCTACCACGTGGACTTCAAACAGAAGCCGTATACTCCGAAGGTGTTCAGAGAAGTTACAGTCAAAGGATTCGACGCGTCGCAGTACGAGGCCAAACAGATATTCTACTCCAGTAAAGACGGCACCAAAGTACCCATGTTCATTATTTCGAAAAAGGATCTGCCGCGCGACGGCTCGAACCCGGTCCTGCTGTACGGCTACGGCGGCTTCAACATCAACGTGCAGCCCGGCTTCAGCGTCACGCGGCTCGTCTTCATGCAGCACATGAACGGGATCGTCGCCATCCCCAACATCAGGGGCGGCGGGGAATACGGCGAGAGGTGGCACAACGCGGGGCGGCTGCTGAACAAGCAGAACGTGTTCGACGACTTCCAGGCGGCGGCCGAGTACATGGTGTCGGAGCGGTACACGCGCCCCGCCCTCCTCACCATCCAGGGGGGCTCCAACGGCGGCCTGCTCGTCGCCGCCTGCATCAACCAGCGCCCCGACCTCTACGGGGCCGCCGTCGTGCAGGTCGGAGTTCTTGACATGCTACGCTTCCAGAAGTTCACGATAGGACACGCCTGGGTGTCTGACTACGGAAGCTCTGACAATAAGACCCAGTTCGAGTACTTGCTGAAGTACTCGCCCTTACACAACATTCAGCCGCCGAGCGAGAATCGGCCCGAATACCCGGCGACGCTGATACTGTCGGCGGACCACGACGACCGCGTGGTGCCGCTGCACTCGCTGAAGTTCGTGGCCGAACTGCAGCACGTGGCGGGGCGCTCGCCGGCGCAGCGCGCGCCGCTGCTCGCCAGGTTCGACACCAAGGCGGGGCACGGCGGCGGAAAACCCACCACTAAAATTATTGACGAACACACTGATATCCTATGTTTCATGACGCAAGCTTTGGGTCTGAAGTTTGTAAAATGA
Protein
MGLVFAMRACIICVLIACAFLQCTGTTILSSRNMLYDYPAVRRDETVVDDYHGTKIKDPYRWLEDPDSNETKEFVEAQNKITRPYLDACPVQKSINERLTELWNYPKYSCPFRKGSRYFFFKNTGLQNQNALYVQDGLDGEPRVFLDPNTLSEDGTVALSGTKFTEDGSTLAYGLSASGSDWIAIHFKDVATGEDYPEVLEKVKFASMSWTKDNKGLFYSMYPKQTGKTDGSETEVNHDQKLCYHRINTSQSEDVVVVEFPEEPLWRIVADVSDCGRYLLVYPVRDSRDNLLFFADLSKHPEINGKLPLMPVVEEFEADYEYVTNEDSVCIIRTNKNAPNYRLIMIDLQNPAKENWRTLVPEHPTDVLDWAAAVDQDKLVIHYIRDVKSVLQLHDLKTGEFIQTFPLDVGSVVGFTGKKNQTEIFYHFMSFLTPGVIYHVDFKQKPYTPKVFREVTVKGFDASQYEAKQIFYSSKDGTKVPMFIISKKDLPRDGSNPVLLYGYGGFNINVQPGFSVTRLVFMQHMNGIVAIPNIRGGGEYGERWHNAGRLLNKQNVFDDFQAAAEYMVSERYTRPALLTIQGGSNGGLLVAACINQRPDLYGAAVVQVGVLDMLRFQKFTIGHAWVSDYGSSDNKTQFEYLLKYSPLHNIQPPSENRPEYPATLILSADHDDRVVPLHSLKFVAELQHVAGRSPAQRAPLLARFDTKAGHGGGKPTTKIIDEHTDILCFMTQALGLKFVK
Summary
Uniprot
A0A2U8JEW8
A0A2U8JEW0
A0A2A4JHM7
A0A194QWQ8
A0A2H1X011
A0A2A4JIM1
+ More
A0A2A4JIG0 A0A194PL24 A0A2A4JID3 S4PE14 A0A212FKH5 A0A3S2P7R8 A0A2J7QKX9 A0A2J7QKX5 A0A067RCU9 A0A232F161 K7IRE4 E2AJM5 A0A0C9QHX8 A0A0P4W8Y4 A0A3L8DC24 A0A026WJZ5 A0A1B6DCC9 A0A088AQ94 E2C617 A0A2A3EIC2 A0A195C2T1 A0A151JMH9 A0A0L7QYW9 A0A1I9WLN1 T1IS74 A0A0J7KAW6 A0A154PJE0 A0A182F1M3 A0A2M3ZGC0 A0A2M3Z2G8 A0A2M3Z2B8 W5JJ34 A0A2M4BEQ7 A0A2M4BEU4 A0A2M4BEQ4 A0A084VMW7 A0A2M4A9K0 A0A0D3QDV3 A0A182NF44 A0A195BKS2 A0A182PFR3 A0A0D3QD94 A0A182QTK2 A0A182J255 A0A1S4H140 A0A0D3QDV5 A0A0D3QD76 A0A0D3QDZ9 A0A0D3QDV7 A0A0D3QD98 A0A0D3QD88 A0A1I8Q7E1 A0A182R6J3 Q7PVV8 A0A182UP27 A0A182K9P5 A0A0D3QDK2 A0A0D3QD86 A0A182I560 A0A0D3QE05 A0A182TIV1 A0A182WYL0 A0A182ME33 A0A1I8MQN1 A0A182Y9S0 T1PFY3 A0A1S3II94 Q16WP2 A0A182H044 A0A182WH65 A0A131XSG7 A0A1L8G2Y9 A0A0P5XEX7 A0A1L8G9B4 A0A1Q3F9L0 Q7ZWL1 A0A1Q3F9J6 A0A0P5EA08 Q6P4W3 Q6IR92 A0A1Q3F9B1 A5D7C6 A0A0N8CST3 A0A452F941 A0A0P5BIC5 A0A0P5VA38 B3MJH3 A0A0N8A2N9 A0A0P5WZ65 A0A0P5NHY7 A0A0P6IV69 Q4KLX3 A0A3Q0CQK6 Q543B9
A0A2A4JIG0 A0A194PL24 A0A2A4JID3 S4PE14 A0A212FKH5 A0A3S2P7R8 A0A2J7QKX9 A0A2J7QKX5 A0A067RCU9 A0A232F161 K7IRE4 E2AJM5 A0A0C9QHX8 A0A0P4W8Y4 A0A3L8DC24 A0A026WJZ5 A0A1B6DCC9 A0A088AQ94 E2C617 A0A2A3EIC2 A0A195C2T1 A0A151JMH9 A0A0L7QYW9 A0A1I9WLN1 T1IS74 A0A0J7KAW6 A0A154PJE0 A0A182F1M3 A0A2M3ZGC0 A0A2M3Z2G8 A0A2M3Z2B8 W5JJ34 A0A2M4BEQ7 A0A2M4BEU4 A0A2M4BEQ4 A0A084VMW7 A0A2M4A9K0 A0A0D3QDV3 A0A182NF44 A0A195BKS2 A0A182PFR3 A0A0D3QD94 A0A182QTK2 A0A182J255 A0A1S4H140 A0A0D3QDV5 A0A0D3QD76 A0A0D3QDZ9 A0A0D3QDV7 A0A0D3QD98 A0A0D3QD88 A0A1I8Q7E1 A0A182R6J3 Q7PVV8 A0A182UP27 A0A182K9P5 A0A0D3QDK2 A0A0D3QD86 A0A182I560 A0A0D3QE05 A0A182TIV1 A0A182WYL0 A0A182ME33 A0A1I8MQN1 A0A182Y9S0 T1PFY3 A0A1S3II94 Q16WP2 A0A182H044 A0A182WH65 A0A131XSG7 A0A1L8G2Y9 A0A0P5XEX7 A0A1L8G9B4 A0A1Q3F9L0 Q7ZWL1 A0A1Q3F9J6 A0A0P5EA08 Q6P4W3 Q6IR92 A0A1Q3F9B1 A5D7C6 A0A0N8CST3 A0A452F941 A0A0P5BIC5 A0A0P5VA38 B3MJH3 A0A0N8A2N9 A0A0P5WZ65 A0A0P5NHY7 A0A0P6IV69 Q4KLX3 A0A3Q0CQK6 Q543B9
Pubmed
29753046
26354079
23622113
22118469
24845553
28648823
+ More
20075255 20798317 30249741 24508170 27538518 20920257 23761445 24438588 25552603 12364791 25315136 25244985 17510324 26483478 27762356 19393038 17994087 26319212 28071753 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079
20075255 20798317 30249741 24508170 27538518 20920257 23761445 24438588 25552603 12364791 25315136 25244985 17510324 26483478 27762356 19393038 17994087 26319212 28071753 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079
EMBL
MF538582
AWK60412.1
MF538581
AWK60411.1
NWSH01001356
PCG71557.1
+ More
KQ461103 KPJ09395.1 ODYU01011939 SOQ57974.1 PCG71558.1 PCG71556.1 KQ459606 KPI91805.1 PCG71559.1 GAIX01003391 JAA89169.1 AGBW02008034 OWR54238.1 RSAL01000228 RVE43909.1 NEVH01013256 PNF29235.1 PNF29234.1 KK852549 KDR21572.1 NNAY01001342 OXU24299.1 GL440049 EFN66352.1 GBYB01014263 GBYB01014264 JAG84030.1 JAG84031.1 GDRN01077074 JAI62799.1 QOIP01000010 RLU17904.1 KK107167 EZA56288.1 GEDC01013954 JAS23344.1 GL452895 EFN76622.1 KZ288232 PBC31448.1 KQ978317 KYM95169.1 KQ978949 KYN27382.1 KQ414685 KOC63824.1 KU932415 APA34051.1 JH431425 LBMM01010506 KMQ87419.1 KQ434936 KZC11942.1 GGFM01006853 MBW27604.1 GGFM01001887 MBW22638.1 GGFM01001905 MBW22656.1 ADMH02001355 ETN62890.1 GGFJ01002394 MBW51535.1 GGFJ01002396 MBW51537.1 GGFJ01002395 MBW51536.1 ATLV01014632 KE524975 KFB39311.1 GGFK01004091 MBW37412.1 KP274684 AJC98850.1 KQ976453 KYM85369.1 KP274681 KP274690 AJC98847.1 AJC98856.1 AXCN02000829 AAAB01008984 KP274689 AJC98855.1 KP274679 KP274682 KP274686 KP274687 KP274691 KP274693 AJC98845.1 AJC98848.1 AJC98852.1 AJC98853.1 AJC98857.1 AJC98859.1 KP274680 KP274688 AJC98846.1 AJC98854.1 KP274694 AJC98860.1 KP274695 AJC98861.1 KP274683 KP274685 KP274692 AJC98849.1 AJC98851.1 AJC98858.1 EAA14977.4 KP274696 AJC98862.1 KP274697 AJC98863.1 APCN01003786 KP274698 AJC98864.1 AXCM01001716 KA647666 AFP62295.1 CH477557 EAT39025.1 JXUM01100695 JXUM01100696 JXUM01100697 KQ564590 KXJ72049.1 GEFM01006514 JAP69282.1 CM004475 OCT78208.1 GDIP01073048 JAM30667.1 CM004474 OCT80423.1 GFDL01010842 JAV24203.1 BC047161 AAH47161.1 GFDL01010794 JAV24251.1 GDIP01203968 GDIP01146127 GDIP01035653 LRGB01002384 JAJ77275.1 KZS07862.1 BC063222 AAH63222.1 BC071008 AAH71008.1 GFDL01010895 JAV24150.1 BC140507 AAI40508.1 GDIP01102383 JAM01332.1 LWLT01000012 GDIP01188426 JAJ34976.1 GDIP01102385 JAM01330.1 CH902620 EDV32341.1 GDIP01188425 JAJ34977.1 GDIP01079483 JAM24232.1 GDIQ01152420 JAK99305.1 GDIQ01004374 JAN90363.1 BC098959 AAH98959.1 AK053816 AK141851 AK161528 AK163787 CH466540 BAC35538.1 BAE24858.1 BAE36445.1 BAE37495.1 EDL05036.1
KQ461103 KPJ09395.1 ODYU01011939 SOQ57974.1 PCG71558.1 PCG71556.1 KQ459606 KPI91805.1 PCG71559.1 GAIX01003391 JAA89169.1 AGBW02008034 OWR54238.1 RSAL01000228 RVE43909.1 NEVH01013256 PNF29235.1 PNF29234.1 KK852549 KDR21572.1 NNAY01001342 OXU24299.1 GL440049 EFN66352.1 GBYB01014263 GBYB01014264 JAG84030.1 JAG84031.1 GDRN01077074 JAI62799.1 QOIP01000010 RLU17904.1 KK107167 EZA56288.1 GEDC01013954 JAS23344.1 GL452895 EFN76622.1 KZ288232 PBC31448.1 KQ978317 KYM95169.1 KQ978949 KYN27382.1 KQ414685 KOC63824.1 KU932415 APA34051.1 JH431425 LBMM01010506 KMQ87419.1 KQ434936 KZC11942.1 GGFM01006853 MBW27604.1 GGFM01001887 MBW22638.1 GGFM01001905 MBW22656.1 ADMH02001355 ETN62890.1 GGFJ01002394 MBW51535.1 GGFJ01002396 MBW51537.1 GGFJ01002395 MBW51536.1 ATLV01014632 KE524975 KFB39311.1 GGFK01004091 MBW37412.1 KP274684 AJC98850.1 KQ976453 KYM85369.1 KP274681 KP274690 AJC98847.1 AJC98856.1 AXCN02000829 AAAB01008984 KP274689 AJC98855.1 KP274679 KP274682 KP274686 KP274687 KP274691 KP274693 AJC98845.1 AJC98848.1 AJC98852.1 AJC98853.1 AJC98857.1 AJC98859.1 KP274680 KP274688 AJC98846.1 AJC98854.1 KP274694 AJC98860.1 KP274695 AJC98861.1 KP274683 KP274685 KP274692 AJC98849.1 AJC98851.1 AJC98858.1 EAA14977.4 KP274696 AJC98862.1 KP274697 AJC98863.1 APCN01003786 KP274698 AJC98864.1 AXCM01001716 KA647666 AFP62295.1 CH477557 EAT39025.1 JXUM01100695 JXUM01100696 JXUM01100697 KQ564590 KXJ72049.1 GEFM01006514 JAP69282.1 CM004475 OCT78208.1 GDIP01073048 JAM30667.1 CM004474 OCT80423.1 GFDL01010842 JAV24203.1 BC047161 AAH47161.1 GFDL01010794 JAV24251.1 GDIP01203968 GDIP01146127 GDIP01035653 LRGB01002384 JAJ77275.1 KZS07862.1 BC063222 AAH63222.1 BC071008 AAH71008.1 GFDL01010895 JAV24150.1 BC140507 AAI40508.1 GDIP01102383 JAM01332.1 LWLT01000012 GDIP01188426 JAJ34976.1 GDIP01102385 JAM01330.1 CH902620 EDV32341.1 GDIP01188425 JAJ34977.1 GDIP01079483 JAM24232.1 GDIQ01152420 JAK99305.1 GDIQ01004374 JAN90363.1 BC098959 AAH98959.1 AK053816 AK141851 AK161528 AK163787 CH466540 BAC35538.1 BAE24858.1 BAE36445.1 BAE37495.1 EDL05036.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
UP000235965
+ More
UP000027135 UP000215335 UP000002358 UP000000311 UP000279307 UP000053097 UP000005203 UP000008237 UP000242457 UP000078542 UP000078492 UP000053825 UP000036403 UP000076502 UP000069272 UP000000673 UP000030765 UP000075884 UP000078540 UP000075885 UP000075886 UP000075880 UP000095300 UP000075900 UP000007062 UP000075903 UP000075881 UP000075840 UP000075902 UP000076407 UP000075883 UP000095301 UP000076408 UP000085678 UP000008820 UP000069940 UP000249989 UP000075920 UP000186698 UP000076858 UP000009136 UP000291000 UP000007801 UP000189706
UP000027135 UP000215335 UP000002358 UP000000311 UP000279307 UP000053097 UP000005203 UP000008237 UP000242457 UP000078542 UP000078492 UP000053825 UP000036403 UP000076502 UP000069272 UP000000673 UP000030765 UP000075884 UP000078540 UP000075885 UP000075886 UP000075880 UP000095300 UP000075900 UP000007062 UP000075903 UP000075881 UP000075840 UP000075902 UP000076407 UP000075883 UP000095301 UP000076408 UP000085678 UP000008820 UP000069940 UP000249989 UP000075920 UP000186698 UP000076858 UP000009136 UP000291000 UP000007801 UP000189706
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2U8JEW8
A0A2U8JEW0
A0A2A4JHM7
A0A194QWQ8
A0A2H1X011
A0A2A4JIM1
+ More
A0A2A4JIG0 A0A194PL24 A0A2A4JID3 S4PE14 A0A212FKH5 A0A3S2P7R8 A0A2J7QKX9 A0A2J7QKX5 A0A067RCU9 A0A232F161 K7IRE4 E2AJM5 A0A0C9QHX8 A0A0P4W8Y4 A0A3L8DC24 A0A026WJZ5 A0A1B6DCC9 A0A088AQ94 E2C617 A0A2A3EIC2 A0A195C2T1 A0A151JMH9 A0A0L7QYW9 A0A1I9WLN1 T1IS74 A0A0J7KAW6 A0A154PJE0 A0A182F1M3 A0A2M3ZGC0 A0A2M3Z2G8 A0A2M3Z2B8 W5JJ34 A0A2M4BEQ7 A0A2M4BEU4 A0A2M4BEQ4 A0A084VMW7 A0A2M4A9K0 A0A0D3QDV3 A0A182NF44 A0A195BKS2 A0A182PFR3 A0A0D3QD94 A0A182QTK2 A0A182J255 A0A1S4H140 A0A0D3QDV5 A0A0D3QD76 A0A0D3QDZ9 A0A0D3QDV7 A0A0D3QD98 A0A0D3QD88 A0A1I8Q7E1 A0A182R6J3 Q7PVV8 A0A182UP27 A0A182K9P5 A0A0D3QDK2 A0A0D3QD86 A0A182I560 A0A0D3QE05 A0A182TIV1 A0A182WYL0 A0A182ME33 A0A1I8MQN1 A0A182Y9S0 T1PFY3 A0A1S3II94 Q16WP2 A0A182H044 A0A182WH65 A0A131XSG7 A0A1L8G2Y9 A0A0P5XEX7 A0A1L8G9B4 A0A1Q3F9L0 Q7ZWL1 A0A1Q3F9J6 A0A0P5EA08 Q6P4W3 Q6IR92 A0A1Q3F9B1 A5D7C6 A0A0N8CST3 A0A452F941 A0A0P5BIC5 A0A0P5VA38 B3MJH3 A0A0N8A2N9 A0A0P5WZ65 A0A0P5NHY7 A0A0P6IV69 Q4KLX3 A0A3Q0CQK6 Q543B9
A0A2A4JIG0 A0A194PL24 A0A2A4JID3 S4PE14 A0A212FKH5 A0A3S2P7R8 A0A2J7QKX9 A0A2J7QKX5 A0A067RCU9 A0A232F161 K7IRE4 E2AJM5 A0A0C9QHX8 A0A0P4W8Y4 A0A3L8DC24 A0A026WJZ5 A0A1B6DCC9 A0A088AQ94 E2C617 A0A2A3EIC2 A0A195C2T1 A0A151JMH9 A0A0L7QYW9 A0A1I9WLN1 T1IS74 A0A0J7KAW6 A0A154PJE0 A0A182F1M3 A0A2M3ZGC0 A0A2M3Z2G8 A0A2M3Z2B8 W5JJ34 A0A2M4BEQ7 A0A2M4BEU4 A0A2M4BEQ4 A0A084VMW7 A0A2M4A9K0 A0A0D3QDV3 A0A182NF44 A0A195BKS2 A0A182PFR3 A0A0D3QD94 A0A182QTK2 A0A182J255 A0A1S4H140 A0A0D3QDV5 A0A0D3QD76 A0A0D3QDZ9 A0A0D3QDV7 A0A0D3QD98 A0A0D3QD88 A0A1I8Q7E1 A0A182R6J3 Q7PVV8 A0A182UP27 A0A182K9P5 A0A0D3QDK2 A0A0D3QD86 A0A182I560 A0A0D3QE05 A0A182TIV1 A0A182WYL0 A0A182ME33 A0A1I8MQN1 A0A182Y9S0 T1PFY3 A0A1S3II94 Q16WP2 A0A182H044 A0A182WH65 A0A131XSG7 A0A1L8G2Y9 A0A0P5XEX7 A0A1L8G9B4 A0A1Q3F9L0 Q7ZWL1 A0A1Q3F9J6 A0A0P5EA08 Q6P4W3 Q6IR92 A0A1Q3F9B1 A5D7C6 A0A0N8CST3 A0A452F941 A0A0P5BIC5 A0A0P5VA38 B3MJH3 A0A0N8A2N9 A0A0P5WZ65 A0A0P5NHY7 A0A0P6IV69 Q4KLX3 A0A3Q0CQK6 Q543B9
PDB
3DDU
E-value=0,
Score=2311
Ontologies
GO
Topology
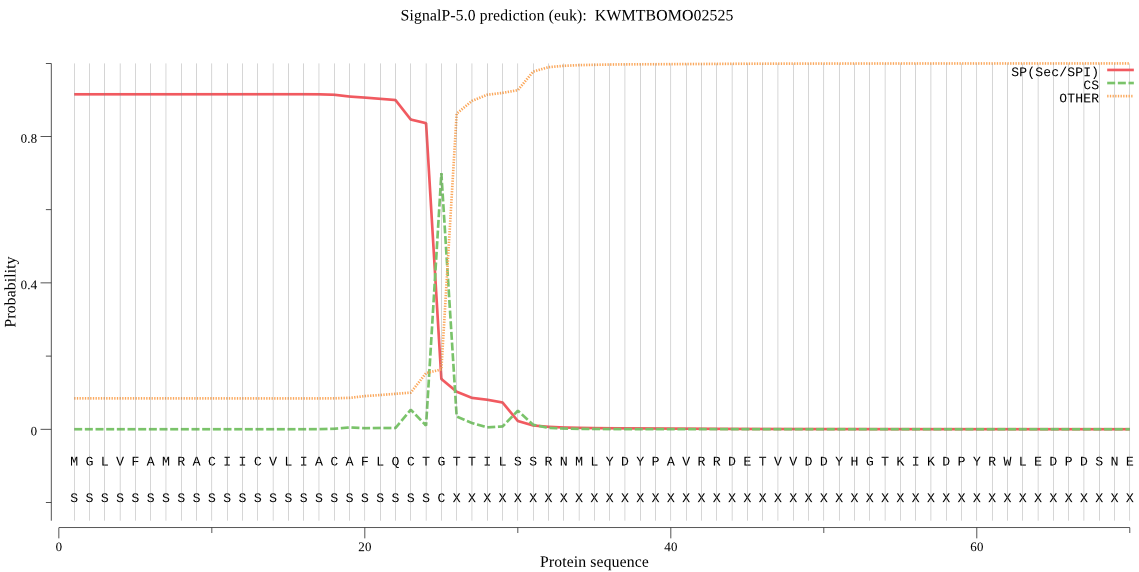
SignalP
Position: 1 - 25,
Likelihood: 0.915978
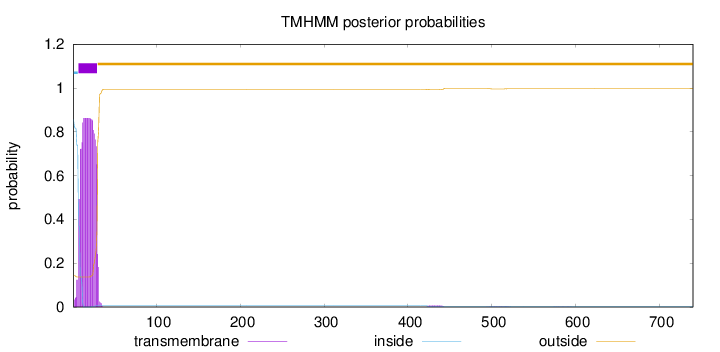
Length:
740
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.1430199999999
Exp number, first 60 AAs:
18.96209
Total prob of N-in:
0.85726
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 740
Population Genetic Test Statistics
Pi
245.425704
Theta
176.851376
Tajima's D
1.076251
CLR
1.11143
CSRT
0.683265836708165
Interpretation
Uncertain
