Gene
KWMTBOMO02518
Pre Gene Modal
BGIBMGA002696
Annotation
PREDICTED:_uncharacterized_protein_LOC101740429_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.819
Sequence
CDS
ATGACCCCGTCTCGATACACGCCAAGACCTACGAGTACACGTGGGCGCGGTTCAGCGCATTTCTCTACGTCGAGTGGATCAGATGCACCTCTGCCGCCACCCAACCGAGGAACACCCCCAACACGCAGCCGGCCGACATTGAAACCCTCCACAGCTATTGTCTCAAAAACGACAGAGTTTGTAGACATCTACAACCATCCGCCCAGTAGACCAGCGTCTGTTTATCCTCAACCAACACCGGATAAGACAGCGGCGAAATGCAGGAAAGACGTTTGTTTGTTGCCAGACTGCTACTGCGGTGGAAAGGAAATTCCAGGAGACTTACCGGTGGAGTCGGTCCCGCAAATTGTTCTTCTGACATTTGACGATTCCGTCAACGATTTGAACAAAGGCCTGTATACTGACCTCTTTGAGAAAGGTCGTGTCAACCCGAACGGCTGTCCGATTACGGCAACATTTTATGTCTCCCACGAATGGACTGATTACAGCCAAGTTCAGAACCTCTATTCTGCTGGACACGAGATGGCCTCACACACGATTTCCCATAGCTTTGGTGAACAATTTTCTCAAAAGAAATGGAACAGAGAAGTTGCTGGCCAACGAGAAATTCTTGCAGCTTATGGTGGTGTAAAACTTGAGGATGTTAGAGGAATGAGAGCGCCGTTCCTTTCCGTTGGTGGAAACAAAATGTTCAAAATGCTGTACGATTCTAACTTCACTTACGACTCTTCTCTCCCAGTGTATGAAAACAGACCGCCCAGCTGGCCTTATACTCTCGATTATAAACTATTCCACGACTGTATGATACCACCGTGTCCGACGAAATCCTATCCAGGTGTTTGGGAGGTCCCCATGGTGATGTGGCAAGACTTGAACGGGGGCCGATGCTCCATGGGTGATGCTTGCGCTAATCCACCTGAACCCGAAAACGTTTATAAAATGATTCTGAAGAACTTCGATAGACACTATAGCACCAACAGGGCACCTTTCGGTTTATTCTATCACGCGGCTTGGTTCACTCAGCCGCACCACAAGGAAGGCTTCATTATGTTCTTGGATCACATCAACCAGATGCCGGATGTGTGGATAGTCACTAACTGGCAAGCTCTACAGTGGGTCAGGGATCCCACTCCTATCTCTAGGATAAATAATTTCCAGCCTTTCCAGTGCAACTATCAGGATCGACCGAAGAAGTGCAACAACCCCAAAGTGTGCAACCTTTGGCACAAATCCGGAGTAAGATACATGAGAACATGTCAACCCTGTCCCGAAGTGTACCCATGGACCGGCAAACAGTCGGAGAGCAAAGTTGTTGTAGTCGCTAGATCTTCGCAATGGGCGGTACATGAATTCTTATCTAGTTCATTTTCAAGAGGATTTATCAATTTAGTAGTCATCGGACAGAGTTTCAAAGAAGACGATGATTCAACTATAGAATCACCGTATATATTATACACTCATAAATTATACACGGACGGATTGGGTGCAAGTAAGCCTGTCGTTCTCAATTCTTGGTCTCATGGAAAGTTTTCGAGGAATGTAAATTTGTTTCCACCAAAAATGACAGGAGGGTATGCGGGACATAGAGTCGTCGTAGCTGCAGCCAATCAACCACCTTTCGTTTTTAGAAGGTAA
Protein
MTPSRYTPRPTSTRGRGSAHFSTSSGSDAPLPPPNRGTPPTRSRPTLKPSTAIVSKTTEFVDIYNHPPSRPASVYPQPTPDKTAAKCRKDVCLLPDCYCGGKEIPGDLPVESVPQIVLLTFDDSVNDLNKGLYTDLFEKGRVNPNGCPITATFYVSHEWTDYSQVQNLYSAGHEMASHTISHSFGEQFSQKKWNREVAGQREILAAYGGVKLEDVRGMRAPFLSVGGNKMFKMLYDSNFTYDSSLPVYENRPPSWPYTLDYKLFHDCMIPPCPTKSYPGVWEVPMVMWQDLNGGRCSMGDACANPPEPENVYKMILKNFDRHYSTNRAPFGLFYHAAWFTQPHHKEGFIMFLDHINQMPDVWIVTNWQALQWVRDPTPISRINNFQPFQCNYQDRPKKCNNPKVCNLWHKSGVRYMRTCQPCPEVYPWTGKQSESKVVVVARSSQWAVHEFLSSSFSRGFINLVVIGQSFKEDDDSTIESPYILYTHKLYTDGLGASKPVVLNSWSHGKFSRNVNLFPPKMTGGYAGHRVVVAAANQPPFVFRR
Summary
Uniprot
H9IZL1
A0A2H1WFW7
A0A212FCK4
I4DRF5
A0A0K8U465
A0A034VP01
+ More
B4MDB2 A0A1W4V2W0 B3MLU1 A0A1W4V2I8 A0A182RCA8 A0A0P8ZEF2 A0A0P8XUQ7 A0A0A1XH40 A0A1W4UQL4 A0A182JUE9 A0A0P8Y308 A0A182PFA1 A0A0Q9X323 A0A182F2N0 A0A0J9QSX1 A0A182VTQ9 A0A0J9QTR4 A0A182NB34 B3N763 W5JFF6 A0A0J9QT37 A0A1S4FZ63 A0A3B0JG43 A0A0J9QSQ9 A0A0J9QTS7 A0A182Y8G0 A0A1I8NLI3 A0A1W4V2W5 Q16JD9 A0A1I8MIK2 A0A0Q9XDX6 Q29NQ4 Q9VPI3 A0A0K8U8P0 B4GQS5 A0A0J9TC58 W8C4G8 M9NCX5 A0A1W4V403 A0A0R3NRL1 A0A084VNM0 A0A0J9QT27 A0A0J9TC76 A0A0R3NY10 B4P1U3 A8DYS5 M9NCL3 A0A182X175 A0A1S4GYH8 A0A182IBV3 A0A0N8NZ14 A0A0J9QSX8 A0A0J9QSQ2 Q7Q4P5 A0A0J9QSR7 B0XFK6 A0A0J9QT32 M9NEJ4 A0A182U5N9 A0A2J7PSJ9 A0A1W4UQ32 A0A2J7PSN2 A0A0Q5W8Z3 A0A0K8V5C5 A0A182Q9Q8 A0A2M4CVJ2 A0A2M3ZF46 A0A0P8XEB6 A0A2M3ZDZ4 A0A2M4CVG3 B4ICQ9 A0A0A1X320 A0A2M4AGA2 A0A2M4AGA6 A0A0J9QTS3 A0A0K8SHL6 A0A0A9Y9W0 Q9VPI4 A0A1L8E3V6 A0A1L8DBX9 A0A1L8E3I2 A0A0Q9X349 A0A0R3NRG4 A0A1W4V2J6 A0A0R3NYT3 D7GY93 A8W491 A0A2J7PSK7 A0A182L046 A0A0Q5WK77 A0A0J9TC70 A0A0L0BLP1 A0A0R3NX86 M9NE36
B4MDB2 A0A1W4V2W0 B3MLU1 A0A1W4V2I8 A0A182RCA8 A0A0P8ZEF2 A0A0P8XUQ7 A0A0A1XH40 A0A1W4UQL4 A0A182JUE9 A0A0P8Y308 A0A182PFA1 A0A0Q9X323 A0A182F2N0 A0A0J9QSX1 A0A182VTQ9 A0A0J9QTR4 A0A182NB34 B3N763 W5JFF6 A0A0J9QT37 A0A1S4FZ63 A0A3B0JG43 A0A0J9QSQ9 A0A0J9QTS7 A0A182Y8G0 A0A1I8NLI3 A0A1W4V2W5 Q16JD9 A0A1I8MIK2 A0A0Q9XDX6 Q29NQ4 Q9VPI3 A0A0K8U8P0 B4GQS5 A0A0J9TC58 W8C4G8 M9NCX5 A0A1W4V403 A0A0R3NRL1 A0A084VNM0 A0A0J9QT27 A0A0J9TC76 A0A0R3NY10 B4P1U3 A8DYS5 M9NCL3 A0A182X175 A0A1S4GYH8 A0A182IBV3 A0A0N8NZ14 A0A0J9QSX8 A0A0J9QSQ2 Q7Q4P5 A0A0J9QSR7 B0XFK6 A0A0J9QT32 M9NEJ4 A0A182U5N9 A0A2J7PSJ9 A0A1W4UQ32 A0A2J7PSN2 A0A0Q5W8Z3 A0A0K8V5C5 A0A182Q9Q8 A0A2M4CVJ2 A0A2M3ZF46 A0A0P8XEB6 A0A2M3ZDZ4 A0A2M4CVG3 B4ICQ9 A0A0A1X320 A0A2M4AGA2 A0A2M4AGA6 A0A0J9QTS3 A0A0K8SHL6 A0A0A9Y9W0 Q9VPI4 A0A1L8E3V6 A0A1L8DBX9 A0A1L8E3I2 A0A0Q9X349 A0A0R3NRG4 A0A1W4V2J6 A0A0R3NYT3 D7GY93 A8W491 A0A2J7PSK7 A0A182L046 A0A0Q5WK77 A0A0J9TC70 A0A0L0BLP1 A0A0R3NX86 M9NE36
Pubmed
EMBL
BABH01014405
ODYU01008372
SOQ51907.1
AGBW02009170
OWR51476.1
AK405096
+ More
BAM20495.1 GDHF01030961 JAI21353.1 GAKP01015114 GAKP01015110 JAC43842.1 CH940661 EDW71173.2 CH902620 EDV30812.2 KPU73050.1 KPU73051.1 GBXI01003578 JAD10714.1 KPU73048.1 CH964182 KRF99254.1 CM002910 KMY87198.1 KMY87199.1 KMY87204.1 KMY87213.1 CH954177 EDV57170.2 ADMH02001646 ETN61615.1 KMY87216.1 OUUW01000004 SPP79212.1 KMY87207.1 KMY87214.1 CH478013 EAT34386.1 CH933807 KRG02465.1 CH379059 EAL34164.2 AE014134 AAF51568.3 GDHF01029609 JAI22705.1 CH479187 EDW39947.1 KMY87200.1 GAMC01009606 JAB96949.1 AFH03485.1 KRT03700.1 ATLV01014763 KE524984 KFB39564.1 KMY87206.1 KMY87215.1 KRT03699.1 CM000157 EDW87079.2 ABV53594.2 AFH03483.1 AAAB01008964 APCN01000747 KPU73052.1 KMY87203.1 KMY87202.1 EAA12207.4 KMY87205.1 KMY87212.1 DS232932 EDS26869.1 KMY87211.1 AFH03482.1 NEVH01021927 PNF19315.1 PNF19316.1 KQS69892.1 GDHF01018544 JAI33770.1 AXCN02000644 GGFL01005111 MBW69289.1 GGFM01006436 MBW27187.1 KPU73053.1 GGFM01005972 MBW26723.1 GGFL01005081 MBW69259.1 CH480829 EDW45335.1 GBXI01008775 JAD05517.1 GGFK01006485 MBW39806.1 GGFK01006500 MBW39821.1 KMY87209.1 GBRD01013047 GBRD01013045 JAG52781.1 GBHO01014645 JAG28959.1 BT057995 AAF51567.2 ACM16705.1 GFDF01000869 JAV13215.1 GFDF01010113 JAV03971.1 GFDF01000868 JAV13216.1 KRG02463.1 KRT03703.1 KRT03702.1 KQ971307 EFA13323.2 EU190488 ABW74148.1 PNF19314.1 KQS69890.1 KMY87210.1 JRES01001684 KNC20966.1 KRT03701.1 AFH03484.1
BAM20495.1 GDHF01030961 JAI21353.1 GAKP01015114 GAKP01015110 JAC43842.1 CH940661 EDW71173.2 CH902620 EDV30812.2 KPU73050.1 KPU73051.1 GBXI01003578 JAD10714.1 KPU73048.1 CH964182 KRF99254.1 CM002910 KMY87198.1 KMY87199.1 KMY87204.1 KMY87213.1 CH954177 EDV57170.2 ADMH02001646 ETN61615.1 KMY87216.1 OUUW01000004 SPP79212.1 KMY87207.1 KMY87214.1 CH478013 EAT34386.1 CH933807 KRG02465.1 CH379059 EAL34164.2 AE014134 AAF51568.3 GDHF01029609 JAI22705.1 CH479187 EDW39947.1 KMY87200.1 GAMC01009606 JAB96949.1 AFH03485.1 KRT03700.1 ATLV01014763 KE524984 KFB39564.1 KMY87206.1 KMY87215.1 KRT03699.1 CM000157 EDW87079.2 ABV53594.2 AFH03483.1 AAAB01008964 APCN01000747 KPU73052.1 KMY87203.1 KMY87202.1 EAA12207.4 KMY87205.1 KMY87212.1 DS232932 EDS26869.1 KMY87211.1 AFH03482.1 NEVH01021927 PNF19315.1 PNF19316.1 KQS69892.1 GDHF01018544 JAI33770.1 AXCN02000644 GGFL01005111 MBW69289.1 GGFM01006436 MBW27187.1 KPU73053.1 GGFM01005972 MBW26723.1 GGFL01005081 MBW69259.1 CH480829 EDW45335.1 GBXI01008775 JAD05517.1 GGFK01006485 MBW39806.1 GGFK01006500 MBW39821.1 KMY87209.1 GBRD01013047 GBRD01013045 JAG52781.1 GBHO01014645 JAG28959.1 BT057995 AAF51567.2 ACM16705.1 GFDF01000869 JAV13215.1 GFDF01010113 JAV03971.1 GFDF01000868 JAV13216.1 KRG02463.1 KRT03703.1 KRT03702.1 KQ971307 EFA13323.2 EU190488 ABW74148.1 PNF19314.1 KQS69890.1 KMY87210.1 JRES01001684 KNC20966.1 KRT03701.1 AFH03484.1
Proteomes
UP000005204
UP000007151
UP000008792
UP000192221
UP000007801
UP000075900
+ More
UP000075881 UP000075885 UP000007798 UP000069272 UP000075920 UP000075884 UP000008711 UP000000673 UP000268350 UP000076408 UP000095300 UP000008820 UP000095301 UP000009192 UP000001819 UP000000803 UP000008744 UP000030765 UP000002282 UP000076407 UP000075840 UP000007062 UP000002320 UP000075902 UP000235965 UP000075886 UP000001292 UP000007266 UP000075882 UP000037069
UP000075881 UP000075885 UP000007798 UP000069272 UP000075920 UP000075884 UP000008711 UP000000673 UP000268350 UP000076408 UP000095300 UP000008820 UP000095301 UP000009192 UP000001819 UP000000803 UP000008744 UP000030765 UP000002282 UP000076407 UP000075840 UP000007062 UP000002320 UP000075902 UP000235965 UP000075886 UP000001292 UP000007266 UP000075882 UP000037069
Interpro
ProteinModelPortal
H9IZL1
A0A2H1WFW7
A0A212FCK4
I4DRF5
A0A0K8U465
A0A034VP01
+ More
B4MDB2 A0A1W4V2W0 B3MLU1 A0A1W4V2I8 A0A182RCA8 A0A0P8ZEF2 A0A0P8XUQ7 A0A0A1XH40 A0A1W4UQL4 A0A182JUE9 A0A0P8Y308 A0A182PFA1 A0A0Q9X323 A0A182F2N0 A0A0J9QSX1 A0A182VTQ9 A0A0J9QTR4 A0A182NB34 B3N763 W5JFF6 A0A0J9QT37 A0A1S4FZ63 A0A3B0JG43 A0A0J9QSQ9 A0A0J9QTS7 A0A182Y8G0 A0A1I8NLI3 A0A1W4V2W5 Q16JD9 A0A1I8MIK2 A0A0Q9XDX6 Q29NQ4 Q9VPI3 A0A0K8U8P0 B4GQS5 A0A0J9TC58 W8C4G8 M9NCX5 A0A1W4V403 A0A0R3NRL1 A0A084VNM0 A0A0J9QT27 A0A0J9TC76 A0A0R3NY10 B4P1U3 A8DYS5 M9NCL3 A0A182X175 A0A1S4GYH8 A0A182IBV3 A0A0N8NZ14 A0A0J9QSX8 A0A0J9QSQ2 Q7Q4P5 A0A0J9QSR7 B0XFK6 A0A0J9QT32 M9NEJ4 A0A182U5N9 A0A2J7PSJ9 A0A1W4UQ32 A0A2J7PSN2 A0A0Q5W8Z3 A0A0K8V5C5 A0A182Q9Q8 A0A2M4CVJ2 A0A2M3ZF46 A0A0P8XEB6 A0A2M3ZDZ4 A0A2M4CVG3 B4ICQ9 A0A0A1X320 A0A2M4AGA2 A0A2M4AGA6 A0A0J9QTS3 A0A0K8SHL6 A0A0A9Y9W0 Q9VPI4 A0A1L8E3V6 A0A1L8DBX9 A0A1L8E3I2 A0A0Q9X349 A0A0R3NRG4 A0A1W4V2J6 A0A0R3NYT3 D7GY93 A8W491 A0A2J7PSK7 A0A182L046 A0A0Q5WK77 A0A0J9TC70 A0A0L0BLP1 A0A0R3NX86 M9NE36
B4MDB2 A0A1W4V2W0 B3MLU1 A0A1W4V2I8 A0A182RCA8 A0A0P8ZEF2 A0A0P8XUQ7 A0A0A1XH40 A0A1W4UQL4 A0A182JUE9 A0A0P8Y308 A0A182PFA1 A0A0Q9X323 A0A182F2N0 A0A0J9QSX1 A0A182VTQ9 A0A0J9QTR4 A0A182NB34 B3N763 W5JFF6 A0A0J9QT37 A0A1S4FZ63 A0A3B0JG43 A0A0J9QSQ9 A0A0J9QTS7 A0A182Y8G0 A0A1I8NLI3 A0A1W4V2W5 Q16JD9 A0A1I8MIK2 A0A0Q9XDX6 Q29NQ4 Q9VPI3 A0A0K8U8P0 B4GQS5 A0A0J9TC58 W8C4G8 M9NCX5 A0A1W4V403 A0A0R3NRL1 A0A084VNM0 A0A0J9QT27 A0A0J9TC76 A0A0R3NY10 B4P1U3 A8DYS5 M9NCL3 A0A182X175 A0A1S4GYH8 A0A182IBV3 A0A0N8NZ14 A0A0J9QSX8 A0A0J9QSQ2 Q7Q4P5 A0A0J9QSR7 B0XFK6 A0A0J9QT32 M9NEJ4 A0A182U5N9 A0A2J7PSJ9 A0A1W4UQ32 A0A2J7PSN2 A0A0Q5W8Z3 A0A0K8V5C5 A0A182Q9Q8 A0A2M4CVJ2 A0A2M3ZF46 A0A0P8XEB6 A0A2M3ZDZ4 A0A2M4CVG3 B4ICQ9 A0A0A1X320 A0A2M4AGA2 A0A2M4AGA6 A0A0J9QTS3 A0A0K8SHL6 A0A0A9Y9W0 Q9VPI4 A0A1L8E3V6 A0A1L8DBX9 A0A1L8E3I2 A0A0Q9X349 A0A0R3NRG4 A0A1W4V2J6 A0A0R3NYT3 D7GY93 A8W491 A0A2J7PSK7 A0A182L046 A0A0Q5WK77 A0A0J9TC70 A0A0L0BLP1 A0A0R3NX86 M9NE36
PDB
5Z34
E-value=1.10296e-71,
Score=688
Ontologies
GO
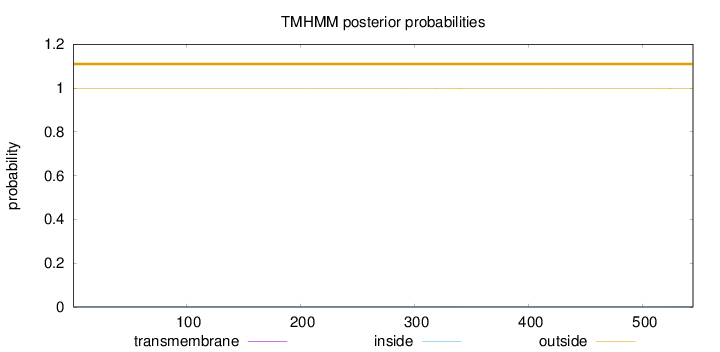
Topology
Length:
544
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01891
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00290
outside
1 - 544
Population Genetic Test Statistics
Pi
185.782432
Theta
168.948854
Tajima's D
0.185486
CLR
130.998746
CSRT
0.424078796060197
Interpretation
Uncertain
