Gene
KWMTBOMO02516
Pre Gene Modal
BGIBMGA002590
Annotation
myocyte_enhancing_factor_2_isoform_B_[Bombyx_mori]
Full name
Myocyte-specific enhancer factor 2
Alternative Name
MADS domain transcription factor
Transcription factor
Location in the cell
Nuclear Reliability : 4.393
Sequence
CDS
ATGGGTCGGAAGAAAATACAAATATCCCGGATTACGGACGAACGGAATCGGCAGGTTACTTTCAACAAACGTAAATTCGGCGTAATGAAGAAGGCGTATGAGTTGAGCGTGCTATGCGACTGTGAGATTGCACTCATCATATTCAGCTCCAACAACAAACTGTACCAATATGCAAGCACGGACATGGACAAGGTTCTGCTGAAATACACGGAATACAATGAACCGCACGAGTCCCTTACGAACCGCAACATCATCGAGAAAGAGCACAAGAACGGAGTGATGTCGCCTGACAGCCCGGAAGCCGAACCCGAATACAACCTGACCCCCCGCACCGAAGCCAAGTACTCGAAGATCGACGAGGAATTCCAAATGATGATGCAAAGGAACCAGTTGAACGGGAGCCGCGTCGGCGTTGGCGTGACGGGAAGCAACTATAACCTACCGGTCAGCGTGCCTGTCGGGAACTACGATCAATCCTTACTCCAAGCTAGTCCACAAATGCATACCTCTATCAGTCCACGGCCATCGTCTTCGGAAACCGATTCAGTATACCCAAGCGGCGGAATGCTCGAAATGTCAAACGGATACCCAGGCTCCGCATCACCACTGGGGGCGGGATGTACCCCGTCGCCGTCCCCCGGCCCCGCTCCCTCTCCCCACCGACACCCGCACAAGTCCCATGCCCACGCTCCGCCACCGCACCACTCGCCCCGACACAACAACCTGCGCGTCGTTATACCTAGTTCAATGCCACCACCGCAAGATGACATATCATATGCAGAGACACCACTAAGCTATTCACTCACCAACTTCGGACCACAGGACTTCAGTATGACCTCGGACATGGGAATCGGACTGACGTGGGGCGCTCACCAACTGCAGACGTTACAACACAACAGTGGCAGCCTGCCGGTATTGGGTGGTGGGGGAACTCCTCCACCGGCCGCGTCACCAAGCAATGTGAAGATCAAAGCGGAACCAGTATCGCCGCCACGCGCACCAGATCACTTGCATCGCGGCCCTCCTCCTGCCAGCGTACCACAGCCCGCACATCAAATGCCAGGAGTCATTGATGGCTCAGTTTCGTCTAGTAACATGGGCTCGCCGGCAGGCCAAGACATGAGGCACTCCAACGCCGTTGCGCTTGACTACGAGCAGCCCCATACCAAGCGGCCGCGGATCGAGGGTTGGGCCACATAG
Protein
MGRKKIQISRITDERNRQVTFNKRKFGVMKKAYELSVLCDCEIALIIFSSNNKLYQYASTDMDKVLLKYTEYNEPHESLTNRNIIEKEHKNGVMSPDSPEAEPEYNLTPRTEAKYSKIDEEFQMMMQRNQLNGSRVGVGVTGSNYNLPVSVPVGNYDQSLLQASPQMHTSISPRPSSSETDSVYPSGGMLEMSNGYPGSASPLGAGCTPSPSPGPAPSPHRHPHKSHAHAPPPHHSPRHNNLRVVIPSSMPPPQDDISYAETPLSYSLTNFGPQDFSMTSDMGIGLTWGAHQLQTLQHNSGSLPVLGGGGTPPPAASPSNVKIKAEPVSPPRAPDHLHRGPPPASVPQPAHQMPGVIDGSVSSSNMGSPAGQDMRHSNAVALDYEQPHTKRPRIEGWAT
Summary
Description
Transcription factor that could be a key player in early mesoderm differentiation and may be required for subsequent cell fate specifications within the somatic and visceral/heart mesodermal layers (PubMed:7839146, PubMed:8052612, PubMed:8202544). Essential for myoblast fusion and consequently muscle formation in adults (PubMed:25797154). During embryonic and pupal development, binds to the enhancer of the myoblast fusion gene sing and activates its transcription (PubMed:25797154).
Similarity
Belongs to the MEF2 family.
Keywords
Activator
Alternative splicing
Complete proteome
Developmental protein
Differentiation
DNA-binding
Myogenesis
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Feature
chain Myocyte-specific enhancer factor 2
splice variant In isoform F.
splice variant In isoform F.
Uniprot
A1E2D3
Q4LG48
A0A2A4IYR6
A0A194R0N3
A0A2H1WFP3
A0A212FCK7
+ More
A0A194PL18 A0A0L7LVI2 A0A1Y1MNF3 A0A1W4X0Q5 A0A1Y1MNE1 A0A1W4WZN4 A0A1W4X0Y4 A0A1Y1MNF0 A0A1W4WPY5 A0A1Y1MNE3 D6W7M3 A0A1L8DIK5 A0A1L8DIM5 A0A1L8DIU4 A0A1L8DIX0 A0A336LL04 U5EYN0 A0A1L8DIL0 A0A0P6K0X9 Q17KU1 A0A1I8MWP6 A0A1I8MWP5 A0A1W4V0M8 A0A1W4UZC4 A0A0R3NP37 A0A1I8MWQ0 B4J5M9 A0A158NSE3 A0A0M4ETH6 A0A1I8MWP7 A0A1I8MWQ1 A0A0A1X2K1 B4LP45 B3MF35 B4GIF6 B0VZE2 A0A0K8W3G2 A0A1W4UM46 A0A1W4UZ20 N6WBY7 A0A1W4V0M3 A0A1I8MWP9 Q28Z04 A0A1I8MWP4 A0A2M4AU80 A0A0A1XT31 P40791-3 A0A1I8MWP3 A0A034W258 P40791-2 A0A0R3NNE9 A0A0A1X5B3 A0A0A9YAJ9 A0A2M4CP32 A0A2M4BJ28 A0A146KQ65 A0A0R3NTW6 A0A0K8UW66 A0A0A9YF65 A0A0K8WC81 A0A087ZQU1 B4MRI7 A0A2A3EK79 A0A0K8WLP4 A0A182XZ71 A0A0B4KFI1 A0A3L8DJI9 A0A2M4AU62 A0A0J7NPK4 A0A2J7QPN5 A0A2M4CP40 A0A2M4BJ05 P40791-4 H9ZYP5 D2WLB9 A0A182KDW2 A0A2M4CPE0
A0A194PL18 A0A0L7LVI2 A0A1Y1MNF3 A0A1W4X0Q5 A0A1Y1MNE1 A0A1W4WZN4 A0A1W4X0Y4 A0A1Y1MNF0 A0A1W4WPY5 A0A1Y1MNE3 D6W7M3 A0A1L8DIK5 A0A1L8DIM5 A0A1L8DIU4 A0A1L8DIX0 A0A336LL04 U5EYN0 A0A1L8DIL0 A0A0P6K0X9 Q17KU1 A0A1I8MWP6 A0A1I8MWP5 A0A1W4V0M8 A0A1W4UZC4 A0A0R3NP37 A0A1I8MWQ0 B4J5M9 A0A158NSE3 A0A0M4ETH6 A0A1I8MWP7 A0A1I8MWQ1 A0A0A1X2K1 B4LP45 B3MF35 B4GIF6 B0VZE2 A0A0K8W3G2 A0A1W4UM46 A0A1W4UZ20 N6WBY7 A0A1W4V0M3 A0A1I8MWP9 Q28Z04 A0A1I8MWP4 A0A2M4AU80 A0A0A1XT31 P40791-3 A0A1I8MWP3 A0A034W258 P40791-2 A0A0R3NNE9 A0A0A1X5B3 A0A0A9YAJ9 A0A2M4CP32 A0A2M4BJ28 A0A146KQ65 A0A0R3NTW6 A0A0K8UW66 A0A0A9YF65 A0A0K8WC81 A0A087ZQU1 B4MRI7 A0A2A3EK79 A0A0K8WLP4 A0A182XZ71 A0A0B4KFI1 A0A3L8DJI9 A0A2M4AU62 A0A0J7NPK4 A0A2J7QPN5 A0A2M4CP40 A0A2M4BJ05 P40791-4 H9ZYP5 D2WLB9 A0A182KDW2 A0A2M4CPE0
Pubmed
16045756
26354079
22118469
26227816
28004739
18362917
+ More
19820115 26999592 17510324 25315136 15632085 17994087 21347285 25830018 8202544 8052612 7605749 7839146 10731132 12537572 12537569 18327897 25797154 25348373 25401762 26823975 25244985 12537568 12537573 12537574 16110336 17569856 17569867 30249741
19820115 26999592 17510324 25315136 15632085 17994087 21347285 25830018 8202544 8052612 7605749 7839146 10731132 12537572 12537569 18327897 25797154 25348373 25401762 26823975 25244985 12537568 12537573 12537574 16110336 17569856 17569867 30249741
EMBL
EF100967
ABL09604.1
AB121093
BAE06225.1
NWSH01004680
PCG64779.1
+ More
KQ461103 KPJ09401.1 ODYU01008372 SOQ51905.1 AGBW02009170 OWR51474.1 KQ459606 KPI91800.1 JTDY01000020 KOB79389.1 GEZM01026166 GEZM01026162 GEZM01026158 JAV87191.1 GEZM01026164 GEZM01026159 GEZM01026157 JAV87193.1 GEZM01026165 GEZM01026161 GEZM01026155 JAV87203.1 GEZM01026163 GEZM01026160 JAV87194.1 KQ971307 EFA11312.2 GFDF01007802 JAV06282.1 GFDF01007803 JAV06281.1 GFDF01007804 JAV06280.1 GFDF01007800 JAV06284.1 UFQS01000040 UFQT01000040 SSW98264.1 SSX18650.1 GANO01000249 JAB59622.1 GFDF01007801 JAV06283.1 GDUN01000553 JAN95366.1 CH477221 EAT47316.1 CM000071 KRT02572.1 CH916367 EDW01805.1 ADTU01024863 CP012524 ALC40557.1 GBXI01008990 JAD05302.1 CH940648 EDW62239.2 CH902619 EDV37663.2 CH479183 EDW36276.1 DS231814 EDS31679.1 GDHF01016901 GDHF01009968 GDHF01006889 JAI35413.1 JAI42346.1 JAI45425.1 ENO01689.2 EAL25811.3 GGFK01011019 MBW44340.1 GBXI01000105 JAD14187.1 U03292 U07422 X83527 U19493 AE013599 AY061096 AY061589 AY118311 GAKP01010193 GAKP01010192 JAC48759.1 KRT02574.1 GBXI01010067 GBXI01008357 GBXI01006137 JAD04225.1 JAD05935.1 JAD08155.1 GBHO01015501 GBHO01015496 GBRD01012754 GBRD01012751 GBRD01012749 JAG28103.1 JAG28108.1 JAG53070.1 GGFL01002902 MBW67080.1 GGFJ01003919 MBW53060.1 GDHC01020440 JAP98188.1 KRT02573.1 GDHF01030477 GDHF01021714 GDHF01018096 JAI21837.1 JAI30600.1 JAI34218.1 GBHO01015504 GBHO01015499 JAG28100.1 JAG28105.1 GDHF01030567 GDHF01003573 JAI21747.1 JAI48741.1 CH963850 EDW74726.2 KZ288226 PBC31894.1 GDHF01026072 GDHF01013320 GDHF01000340 JAI26242.1 JAI38994.1 JAI51974.1 AGB93375.1 QOIP01000007 RLU20333.1 GGFK01010983 MBW44304.1 LBMM01002756 KMQ94410.1 NEVH01012088 PNF30541.1 GGFL01002885 MBW67063.1 GGFJ01003871 MBW53012.1 BT133393 AFH41852.1 GQ386849 GQ386850 ADB24036.1 ADB24037.1 GGFL01002873 MBW67051.1
KQ461103 KPJ09401.1 ODYU01008372 SOQ51905.1 AGBW02009170 OWR51474.1 KQ459606 KPI91800.1 JTDY01000020 KOB79389.1 GEZM01026166 GEZM01026162 GEZM01026158 JAV87191.1 GEZM01026164 GEZM01026159 GEZM01026157 JAV87193.1 GEZM01026165 GEZM01026161 GEZM01026155 JAV87203.1 GEZM01026163 GEZM01026160 JAV87194.1 KQ971307 EFA11312.2 GFDF01007802 JAV06282.1 GFDF01007803 JAV06281.1 GFDF01007804 JAV06280.1 GFDF01007800 JAV06284.1 UFQS01000040 UFQT01000040 SSW98264.1 SSX18650.1 GANO01000249 JAB59622.1 GFDF01007801 JAV06283.1 GDUN01000553 JAN95366.1 CH477221 EAT47316.1 CM000071 KRT02572.1 CH916367 EDW01805.1 ADTU01024863 CP012524 ALC40557.1 GBXI01008990 JAD05302.1 CH940648 EDW62239.2 CH902619 EDV37663.2 CH479183 EDW36276.1 DS231814 EDS31679.1 GDHF01016901 GDHF01009968 GDHF01006889 JAI35413.1 JAI42346.1 JAI45425.1 ENO01689.2 EAL25811.3 GGFK01011019 MBW44340.1 GBXI01000105 JAD14187.1 U03292 U07422 X83527 U19493 AE013599 AY061096 AY061589 AY118311 GAKP01010193 GAKP01010192 JAC48759.1 KRT02574.1 GBXI01010067 GBXI01008357 GBXI01006137 JAD04225.1 JAD05935.1 JAD08155.1 GBHO01015501 GBHO01015496 GBRD01012754 GBRD01012751 GBRD01012749 JAG28103.1 JAG28108.1 JAG53070.1 GGFL01002902 MBW67080.1 GGFJ01003919 MBW53060.1 GDHC01020440 JAP98188.1 KRT02573.1 GDHF01030477 GDHF01021714 GDHF01018096 JAI21837.1 JAI30600.1 JAI34218.1 GBHO01015504 GBHO01015499 JAG28100.1 JAG28105.1 GDHF01030567 GDHF01003573 JAI21747.1 JAI48741.1 CH963850 EDW74726.2 KZ288226 PBC31894.1 GDHF01026072 GDHF01013320 GDHF01000340 JAI26242.1 JAI38994.1 JAI51974.1 AGB93375.1 QOIP01000007 RLU20333.1 GGFK01010983 MBW44304.1 LBMM01002756 KMQ94410.1 NEVH01012088 PNF30541.1 GGFL01002885 MBW67063.1 GGFJ01003871 MBW53012.1 BT133393 AFH41852.1 GQ386849 GQ386850 ADB24036.1 ADB24037.1 GGFL01002873 MBW67051.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
UP000192223
+ More
UP000007266 UP000008820 UP000095301 UP000192221 UP000001819 UP000001070 UP000005205 UP000092553 UP000008792 UP000007801 UP000008744 UP000002320 UP000000803 UP000005203 UP000007798 UP000242457 UP000076408 UP000279307 UP000036403 UP000235965 UP000075881
UP000007266 UP000008820 UP000095301 UP000192221 UP000001819 UP000001070 UP000005205 UP000092553 UP000008792 UP000007801 UP000008744 UP000002320 UP000000803 UP000005203 UP000007798 UP000242457 UP000076408 UP000279307 UP000036403 UP000235965 UP000075881
Interpro
SUPFAM
SSF55455
SSF55455
Gene 3D
ProteinModelPortal
A1E2D3
Q4LG48
A0A2A4IYR6
A0A194R0N3
A0A2H1WFP3
A0A212FCK7
+ More
A0A194PL18 A0A0L7LVI2 A0A1Y1MNF3 A0A1W4X0Q5 A0A1Y1MNE1 A0A1W4WZN4 A0A1W4X0Y4 A0A1Y1MNF0 A0A1W4WPY5 A0A1Y1MNE3 D6W7M3 A0A1L8DIK5 A0A1L8DIM5 A0A1L8DIU4 A0A1L8DIX0 A0A336LL04 U5EYN0 A0A1L8DIL0 A0A0P6K0X9 Q17KU1 A0A1I8MWP6 A0A1I8MWP5 A0A1W4V0M8 A0A1W4UZC4 A0A0R3NP37 A0A1I8MWQ0 B4J5M9 A0A158NSE3 A0A0M4ETH6 A0A1I8MWP7 A0A1I8MWQ1 A0A0A1X2K1 B4LP45 B3MF35 B4GIF6 B0VZE2 A0A0K8W3G2 A0A1W4UM46 A0A1W4UZ20 N6WBY7 A0A1W4V0M3 A0A1I8MWP9 Q28Z04 A0A1I8MWP4 A0A2M4AU80 A0A0A1XT31 P40791-3 A0A1I8MWP3 A0A034W258 P40791-2 A0A0R3NNE9 A0A0A1X5B3 A0A0A9YAJ9 A0A2M4CP32 A0A2M4BJ28 A0A146KQ65 A0A0R3NTW6 A0A0K8UW66 A0A0A9YF65 A0A0K8WC81 A0A087ZQU1 B4MRI7 A0A2A3EK79 A0A0K8WLP4 A0A182XZ71 A0A0B4KFI1 A0A3L8DJI9 A0A2M4AU62 A0A0J7NPK4 A0A2J7QPN5 A0A2M4CP40 A0A2M4BJ05 P40791-4 H9ZYP5 D2WLB9 A0A182KDW2 A0A2M4CPE0
A0A194PL18 A0A0L7LVI2 A0A1Y1MNF3 A0A1W4X0Q5 A0A1Y1MNE1 A0A1W4WZN4 A0A1W4X0Y4 A0A1Y1MNF0 A0A1W4WPY5 A0A1Y1MNE3 D6W7M3 A0A1L8DIK5 A0A1L8DIM5 A0A1L8DIU4 A0A1L8DIX0 A0A336LL04 U5EYN0 A0A1L8DIL0 A0A0P6K0X9 Q17KU1 A0A1I8MWP6 A0A1I8MWP5 A0A1W4V0M8 A0A1W4UZC4 A0A0R3NP37 A0A1I8MWQ0 B4J5M9 A0A158NSE3 A0A0M4ETH6 A0A1I8MWP7 A0A1I8MWQ1 A0A0A1X2K1 B4LP45 B3MF35 B4GIF6 B0VZE2 A0A0K8W3G2 A0A1W4UM46 A0A1W4UZ20 N6WBY7 A0A1W4V0M3 A0A1I8MWP9 Q28Z04 A0A1I8MWP4 A0A2M4AU80 A0A0A1XT31 P40791-3 A0A1I8MWP3 A0A034W258 P40791-2 A0A0R3NNE9 A0A0A1X5B3 A0A0A9YAJ9 A0A2M4CP32 A0A2M4BJ28 A0A146KQ65 A0A0R3NTW6 A0A0K8UW66 A0A0A9YF65 A0A0K8WC81 A0A087ZQU1 B4MRI7 A0A2A3EK79 A0A0K8WLP4 A0A182XZ71 A0A0B4KFI1 A0A3L8DJI9 A0A2M4AU62 A0A0J7NPK4 A0A2J7QPN5 A0A2M4CP40 A0A2M4BJ05 P40791-4 H9ZYP5 D2WLB9 A0A182KDW2 A0A2M4CPE0
PDB
5F28
E-value=1.5113e-40,
Score=418
Ontologies
GO
GO:0045944
GO:0000977
GO:0046983
GO:0005634
GO:0003677
GO:0001228
GO:0048747
GO:0016202
GO:0007507
GO:0007519
GO:0032968
GO:0001158
GO:0019730
GO:0030707
GO:0052576
GO:0007517
GO:0019915
GO:0010468
GO:0007496
GO:0045475
GO:0007520
GO:0007498
GO:0016787
GO:0000165
GO:0006518
GO:0016020
GO:0006631
GO:0006635
GO:0006281
GO:0048384
Topology
Subcellular location
Nucleus
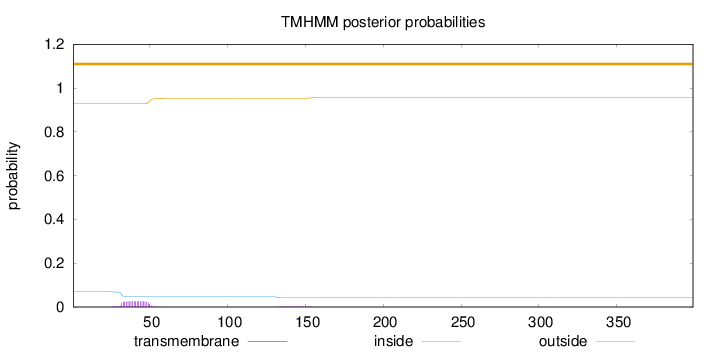
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.57043
Exp number, first 60 AAs:
0.49484
Total prob of N-in:
0.07191
outside
1 - 399
Population Genetic Test Statistics
Pi
210.238838
Theta
176.780025
Tajima's D
0.669578
CLR
0.423471
CSRT
0.570721463926804
Interpretation
Uncertain
