Gene
KWMTBOMO02514 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002587
Annotation
PREDICTED:_peptidylglycine_alpha-hydroxylating_monooxygenase_isoform_X1_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.588
Sequence
CDS
ATGTTTCAATTATTATTTTGTGTTTTTGCCACGGCTGCGGTTTATAATGTTAACGCGTACGAGGTGGACATGTACGAATTTTTAATGCCAAATGTATGGCCGCACTCGGACGAATTGTACCTCTGCACACCAATCAGGATCAGTCCTCGTAACAGATTTAACATAGTCGGCTTTCAACCCAATGCCAGCATGCACACCGCTCACCACATGCTCCTGTATGGATGTTCTGAACCCGGATCTAATGACTCTGTTTGGAATTGCGGTGAAATGCAAAGAAATGTTATCGACGAAATATACAGCACGGCTAGTCCTTGCCGCTCCGGATCACAAATAGTGTACGCTTGGGCACGAGACGCTCCAAGCTTGCATCTTCCAAAAGACGTCGGCTTCCTCGTTGGACAAGGGTCGCCTATAAAATACCTGGTTCTCCAAGTTCATTACATGCATCGATTCCCTAAGGGCCACACGGACAATTCAGGTGTATTCTTGAAGTACACTAAAGCACATATGCCCCGTCAAGCCGGCGTAATACTCCTCGGCACCAGCGGTATAATAATGCCCAACATGGTGGAACACATGGAGACGGCCTGCACCATGACTGAAGACAAGACGATCCACCCGTTCGCTTTCCGTACTCACACGCATTCGCTTGGCACCCTCGTGAGCGGCTACGTCGTCCACCGGAATAAGGACGGAGACGAATGGACATTGCTCGGCAAGAAGAACCCTCAGCTGCCCCAGATGTTCTATCCGACAGAAAACCGAGATCCGATCAAGAAAAATGATGTACTGGCTGCTCGATGCACCATGAATAATAGCCACGAATACGTTGTCAAGATCGGTGCCACAAATCAAGACGAAATGTGCAACTTCTACCTGATGTATTGGGTTGAAAATGACACGCCATTGAAACAGAAGTACTGCTTCACGGCCGGCCCTCCAAACTACTATTGGAGCGAAGCCCACGAGAACTTCAACTGGATCCCGGATCTTGAAGCGAGCACTCCTTAA
Protein
MFQLLFCVFATAAVYNVNAYEVDMYEFLMPNVWPHSDELYLCTPIRISPRNRFNIVGFQPNASMHTAHHMLLYGCSEPGSNDSVWNCGEMQRNVIDEIYSTASPCRSGSQIVYAWARDAPSLHLPKDVGFLVGQGSPIKYLVLQVHYMHRFPKGHTDNSGVFLKYTKAHMPRQAGVILLGTSGIIMPNMVEHMETACTMTEDKTIHPFAFRTHTHSLGTLVSGYVVHRNKDGDEWTLLGKKNPQLPQMFYPTENRDPIKKNDVLAARCTMNNSHEYVVKIGATNQDEMCNFYLMYWVENDTPLKQKYCFTAGPPNYYWSEAHENFNWIPDLEASTP
Summary
Uniprot
H9IZA2
A0A2H1V1X7
A0A194PEU3
A0A212ERW5
A0A2A4JL15
A0A2S1TPB5
+ More
A0A1B6C5R3 A0A1B6JAV9 A0A2P8XPC8 A0A182XYT2 U5EW08 T1PAP5 A0A1B6KT73 A0A1B6EU46 E0W2I3 A0A182IQH4 A0A0L0CB80 A0A1Y1LEP3 A0A182QIJ1 A0A1I8QD09 B0VZG6 A0A1A9X9L2 A0A1B0BQX1 A0A1B0GC47 A0A182KR85 A0A182I379 A0A1Q3FR79 A0A1B0GKM8 A0A2M4CIU5 Q7QJC8 A0A2M3Z726 A0A2M4CIZ7 A0A0P4VMM9 A0A2K8JLU2 A0A2J7R3P3 A0A1L8DTW5 A0A2M4AQX6 A0A1A9WI66 A0A1L8DU00 A0A2M4CJE1 E2C0Z0 A0A2M4BR27 A0A2M3ZHK1 A0A336M3Q7 A0A1D2NN17 A0A2M4APC8 A0A2M4AP62 A0A2M4BP61 A0A182PQI1 A0A0A9WT63 A0A1W6EWE4 A0A3L8DW12 A0A1J1J7B5 A0A034WUI5 A0A0C9R3J0 W8BX66 A0A195CW53 A0A195F777 A0A151J3Z9 A0A0K8WEX9 A0A182KAW2 A0A182FF15 B4LP17 A0A0A1X3W2 A0A1W4X0R0 A0A182X3U2 A0A182V5X6 A0A023EPK4 A0A1S4EYN6 A0A0V0G355 A0A182R7E7 Q17LB1 A0A158NYI4 A0A182MWS9 A0A224XUB0 F4X355 E9IUT2 A0A0L7RDR5 J9JQP7 B4J667 C4WW00 E2A474 A0A2P2I356 A6BL34 A0A023F355 A0A151WQ61 A0A0P5EKF7 A0A026WMJ6 A0A0P5Z712 B4KLW7 A0A067QUJ7 A0A0A9WW54 A0A0P5EIS9 A0A182WJB0 A0A0P5KD98 A0A0P5FB89 A0A164MVI1 A0A0J9U9T9 A0A0P5X007 A0A0P4WEH3
A0A1B6C5R3 A0A1B6JAV9 A0A2P8XPC8 A0A182XYT2 U5EW08 T1PAP5 A0A1B6KT73 A0A1B6EU46 E0W2I3 A0A182IQH4 A0A0L0CB80 A0A1Y1LEP3 A0A182QIJ1 A0A1I8QD09 B0VZG6 A0A1A9X9L2 A0A1B0BQX1 A0A1B0GC47 A0A182KR85 A0A182I379 A0A1Q3FR79 A0A1B0GKM8 A0A2M4CIU5 Q7QJC8 A0A2M3Z726 A0A2M4CIZ7 A0A0P4VMM9 A0A2K8JLU2 A0A2J7R3P3 A0A1L8DTW5 A0A2M4AQX6 A0A1A9WI66 A0A1L8DU00 A0A2M4CJE1 E2C0Z0 A0A2M4BR27 A0A2M3ZHK1 A0A336M3Q7 A0A1D2NN17 A0A2M4APC8 A0A2M4AP62 A0A2M4BP61 A0A182PQI1 A0A0A9WT63 A0A1W6EWE4 A0A3L8DW12 A0A1J1J7B5 A0A034WUI5 A0A0C9R3J0 W8BX66 A0A195CW53 A0A195F777 A0A151J3Z9 A0A0K8WEX9 A0A182KAW2 A0A182FF15 B4LP17 A0A0A1X3W2 A0A1W4X0R0 A0A182X3U2 A0A182V5X6 A0A023EPK4 A0A1S4EYN6 A0A0V0G355 A0A182R7E7 Q17LB1 A0A158NYI4 A0A182MWS9 A0A224XUB0 F4X355 E9IUT2 A0A0L7RDR5 J9JQP7 B4J667 C4WW00 E2A474 A0A2P2I356 A6BL34 A0A023F355 A0A151WQ61 A0A0P5EKF7 A0A026WMJ6 A0A0P5Z712 B4KLW7 A0A067QUJ7 A0A0A9WW54 A0A0P5EIS9 A0A182WJB0 A0A0P5KD98 A0A0P5FB89 A0A164MVI1 A0A0J9U9T9 A0A0P5X007 A0A0P4WEH3
Pubmed
19121390
26354079
22118469
29727440
29403074
25244985
+ More
25315136 20566863 26108605 28004739 20966253 12364791 14747013 17210077 27129103 20798317 27289101 25401762 26823975 30249741 25348373 24495485 17994087 25830018 24945155 17510324 21347285 21719571 21282665 25474469 24508170 24845553 22936249
25315136 20566863 26108605 28004739 20966253 12364791 14747013 17210077 27129103 20798317 27289101 25401762 26823975 30249741 25348373 24495485 17994087 25830018 24945155 17510324 21347285 21719571 21282665 25474469 24508170 24845553 22936249
EMBL
BABH01014418
BABH01014419
BABH01014420
ODYU01000307
SOQ34819.1
KQ459606
+ More
KPI91797.1 AGBW02012906 OWR44227.1 NWSH01001213 PCG72100.1 MF189025 AWI63379.1 GEDC01028481 GEDC01025697 GEDC01015766 GEDC01005460 JAS08817.1 JAS11601.1 JAS21532.1 JAS31838.1 GECU01011394 JAS96312.1 PYGN01001589 PSN33856.1 GANO01003171 JAB56700.1 KA644983 AFP59612.1 GEBQ01025328 JAT14649.1 GECZ01028254 JAS41515.1 DS235878 EEB19839.1 JRES01000644 KNC29673.1 GEZM01057926 JAV72094.1 AXCN02000384 DS231814 EDS31703.1 JXJN01018836 JXJN01018837 CCAG010018989 APCN01000201 GFDL01005010 JAV30035.1 AJWK01030723 AJWK01030724 GGFL01001104 MBW65282.1 AAAB01008807 EAA04612.4 GGFM01003554 MBW24305.1 GGFL01001105 MBW65283.1 GDKW01003012 JAI53583.1 KY031018 ATU82769.1 NEVH01007818 PNF35453.1 GFDF01004196 JAV09888.1 GGFK01009687 MBW43008.1 GFDF01004197 JAV09887.1 GGFL01001103 MBW65281.1 GL451850 EFN78426.1 GGFJ01006379 MBW55520.1 GGFM01007275 MBW28026.1 UFQS01000518 UFQT01000518 SSX04539.1 SSX24902.1 LJIJ01000005 ODN06475.1 GGFK01009302 MBW42623.1 GGFK01009254 MBW42575.1 GGFJ01005661 MBW54802.1 GBHO01031972 GBHO01031970 GDHC01021830 GDHC01018652 GDHC01017493 JAG11632.1 JAG11634.1 JAP96798.1 JAP99976.1 JAQ01136.1 KY563631 ARK20040.1 QOIP01000003 RLU24680.1 CVRI01000074 CRL08280.1 GAKP01000663 JAC58289.1 GBYB01010874 JAG80641.1 GAMC01000655 JAC05901.1 KQ977220 KYN04860.1 KQ981768 KYN35924.1 KQ980228 KYN17160.1 GDHF01002657 JAI49657.1 CH940648 EDW61186.1 GBXI01008964 JAD05328.1 GAPW01002161 JAC11437.1 GECL01003582 JAP02542.1 CH477217 EAT47464.1 ADTU01003909 AXCM01003060 GFTR01004867 JAW11559.1 GL888608 EGI59155.1 GL766050 EFZ15653.1 KQ414613 KOC68998.1 ABLF02017300 CH916367 EDW01925.1 AK341764 BAH72070.1 GL436596 EFN71745.1 IACF01002665 LAB68310.1 AB245092 BAF64529.1 GBBI01002887 JAC15825.1 KQ982843 KYQ50039.1 GDIQ01268957 JAJ82767.1 KK107151 EZA57297.1 GDIP01047764 JAM55951.1 CH933808 EDW09777.1 KK852921 KDR13805.1 GBHO01031973 JAG11631.1 GDIQ01268956 JAJ82768.1 GDIQ01185820 JAK65905.1 GDIQ01265173 JAJ86551.1 LRGB01002937 KZS05403.1 CM002911 KMY96200.1 GDIP01079755 JAM23960.1 GDRN01056990 JAI65756.1
KPI91797.1 AGBW02012906 OWR44227.1 NWSH01001213 PCG72100.1 MF189025 AWI63379.1 GEDC01028481 GEDC01025697 GEDC01015766 GEDC01005460 JAS08817.1 JAS11601.1 JAS21532.1 JAS31838.1 GECU01011394 JAS96312.1 PYGN01001589 PSN33856.1 GANO01003171 JAB56700.1 KA644983 AFP59612.1 GEBQ01025328 JAT14649.1 GECZ01028254 JAS41515.1 DS235878 EEB19839.1 JRES01000644 KNC29673.1 GEZM01057926 JAV72094.1 AXCN02000384 DS231814 EDS31703.1 JXJN01018836 JXJN01018837 CCAG010018989 APCN01000201 GFDL01005010 JAV30035.1 AJWK01030723 AJWK01030724 GGFL01001104 MBW65282.1 AAAB01008807 EAA04612.4 GGFM01003554 MBW24305.1 GGFL01001105 MBW65283.1 GDKW01003012 JAI53583.1 KY031018 ATU82769.1 NEVH01007818 PNF35453.1 GFDF01004196 JAV09888.1 GGFK01009687 MBW43008.1 GFDF01004197 JAV09887.1 GGFL01001103 MBW65281.1 GL451850 EFN78426.1 GGFJ01006379 MBW55520.1 GGFM01007275 MBW28026.1 UFQS01000518 UFQT01000518 SSX04539.1 SSX24902.1 LJIJ01000005 ODN06475.1 GGFK01009302 MBW42623.1 GGFK01009254 MBW42575.1 GGFJ01005661 MBW54802.1 GBHO01031972 GBHO01031970 GDHC01021830 GDHC01018652 GDHC01017493 JAG11632.1 JAG11634.1 JAP96798.1 JAP99976.1 JAQ01136.1 KY563631 ARK20040.1 QOIP01000003 RLU24680.1 CVRI01000074 CRL08280.1 GAKP01000663 JAC58289.1 GBYB01010874 JAG80641.1 GAMC01000655 JAC05901.1 KQ977220 KYN04860.1 KQ981768 KYN35924.1 KQ980228 KYN17160.1 GDHF01002657 JAI49657.1 CH940648 EDW61186.1 GBXI01008964 JAD05328.1 GAPW01002161 JAC11437.1 GECL01003582 JAP02542.1 CH477217 EAT47464.1 ADTU01003909 AXCM01003060 GFTR01004867 JAW11559.1 GL888608 EGI59155.1 GL766050 EFZ15653.1 KQ414613 KOC68998.1 ABLF02017300 CH916367 EDW01925.1 AK341764 BAH72070.1 GL436596 EFN71745.1 IACF01002665 LAB68310.1 AB245092 BAF64529.1 GBBI01002887 JAC15825.1 KQ982843 KYQ50039.1 GDIQ01268957 JAJ82767.1 KK107151 EZA57297.1 GDIP01047764 JAM55951.1 CH933808 EDW09777.1 KK852921 KDR13805.1 GBHO01031973 JAG11631.1 GDIQ01268956 JAJ82768.1 GDIQ01185820 JAK65905.1 GDIQ01265173 JAJ86551.1 LRGB01002937 KZS05403.1 CM002911 KMY96200.1 GDIP01079755 JAM23960.1 GDRN01056990 JAI65756.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000245037
UP000076408
+ More
UP000095301 UP000009046 UP000075880 UP000037069 UP000075886 UP000095300 UP000002320 UP000092443 UP000092460 UP000092444 UP000075882 UP000075840 UP000092461 UP000007062 UP000235965 UP000091820 UP000008237 UP000094527 UP000075885 UP000279307 UP000183832 UP000078542 UP000078541 UP000078492 UP000075881 UP000069272 UP000008792 UP000192223 UP000076407 UP000075903 UP000075900 UP000008820 UP000005205 UP000075883 UP000007755 UP000053825 UP000007819 UP000001070 UP000000311 UP000075809 UP000053097 UP000009192 UP000027135 UP000075920 UP000076858
UP000095301 UP000009046 UP000075880 UP000037069 UP000075886 UP000095300 UP000002320 UP000092443 UP000092460 UP000092444 UP000075882 UP000075840 UP000092461 UP000007062 UP000235965 UP000091820 UP000008237 UP000094527 UP000075885 UP000279307 UP000183832 UP000078542 UP000078541 UP000078492 UP000075881 UP000069272 UP000008792 UP000192223 UP000076407 UP000075903 UP000075900 UP000008820 UP000005205 UP000075883 UP000007755 UP000053825 UP000007819 UP000001070 UP000000311 UP000075809 UP000053097 UP000009192 UP000027135 UP000075920 UP000076858
PRIDE
Interpro
SUPFAM
SSF49742
SSF49742
Gene 3D
ProteinModelPortal
H9IZA2
A0A2H1V1X7
A0A194PEU3
A0A212ERW5
A0A2A4JL15
A0A2S1TPB5
+ More
A0A1B6C5R3 A0A1B6JAV9 A0A2P8XPC8 A0A182XYT2 U5EW08 T1PAP5 A0A1B6KT73 A0A1B6EU46 E0W2I3 A0A182IQH4 A0A0L0CB80 A0A1Y1LEP3 A0A182QIJ1 A0A1I8QD09 B0VZG6 A0A1A9X9L2 A0A1B0BQX1 A0A1B0GC47 A0A182KR85 A0A182I379 A0A1Q3FR79 A0A1B0GKM8 A0A2M4CIU5 Q7QJC8 A0A2M3Z726 A0A2M4CIZ7 A0A0P4VMM9 A0A2K8JLU2 A0A2J7R3P3 A0A1L8DTW5 A0A2M4AQX6 A0A1A9WI66 A0A1L8DU00 A0A2M4CJE1 E2C0Z0 A0A2M4BR27 A0A2M3ZHK1 A0A336M3Q7 A0A1D2NN17 A0A2M4APC8 A0A2M4AP62 A0A2M4BP61 A0A182PQI1 A0A0A9WT63 A0A1W6EWE4 A0A3L8DW12 A0A1J1J7B5 A0A034WUI5 A0A0C9R3J0 W8BX66 A0A195CW53 A0A195F777 A0A151J3Z9 A0A0K8WEX9 A0A182KAW2 A0A182FF15 B4LP17 A0A0A1X3W2 A0A1W4X0R0 A0A182X3U2 A0A182V5X6 A0A023EPK4 A0A1S4EYN6 A0A0V0G355 A0A182R7E7 Q17LB1 A0A158NYI4 A0A182MWS9 A0A224XUB0 F4X355 E9IUT2 A0A0L7RDR5 J9JQP7 B4J667 C4WW00 E2A474 A0A2P2I356 A6BL34 A0A023F355 A0A151WQ61 A0A0P5EKF7 A0A026WMJ6 A0A0P5Z712 B4KLW7 A0A067QUJ7 A0A0A9WW54 A0A0P5EIS9 A0A182WJB0 A0A0P5KD98 A0A0P5FB89 A0A164MVI1 A0A0J9U9T9 A0A0P5X007 A0A0P4WEH3
A0A1B6C5R3 A0A1B6JAV9 A0A2P8XPC8 A0A182XYT2 U5EW08 T1PAP5 A0A1B6KT73 A0A1B6EU46 E0W2I3 A0A182IQH4 A0A0L0CB80 A0A1Y1LEP3 A0A182QIJ1 A0A1I8QD09 B0VZG6 A0A1A9X9L2 A0A1B0BQX1 A0A1B0GC47 A0A182KR85 A0A182I379 A0A1Q3FR79 A0A1B0GKM8 A0A2M4CIU5 Q7QJC8 A0A2M3Z726 A0A2M4CIZ7 A0A0P4VMM9 A0A2K8JLU2 A0A2J7R3P3 A0A1L8DTW5 A0A2M4AQX6 A0A1A9WI66 A0A1L8DU00 A0A2M4CJE1 E2C0Z0 A0A2M4BR27 A0A2M3ZHK1 A0A336M3Q7 A0A1D2NN17 A0A2M4APC8 A0A2M4AP62 A0A2M4BP61 A0A182PQI1 A0A0A9WT63 A0A1W6EWE4 A0A3L8DW12 A0A1J1J7B5 A0A034WUI5 A0A0C9R3J0 W8BX66 A0A195CW53 A0A195F777 A0A151J3Z9 A0A0K8WEX9 A0A182KAW2 A0A182FF15 B4LP17 A0A0A1X3W2 A0A1W4X0R0 A0A182X3U2 A0A182V5X6 A0A023EPK4 A0A1S4EYN6 A0A0V0G355 A0A182R7E7 Q17LB1 A0A158NYI4 A0A182MWS9 A0A224XUB0 F4X355 E9IUT2 A0A0L7RDR5 J9JQP7 B4J667 C4WW00 E2A474 A0A2P2I356 A6BL34 A0A023F355 A0A151WQ61 A0A0P5EKF7 A0A026WMJ6 A0A0P5Z712 B4KLW7 A0A067QUJ7 A0A0A9WW54 A0A0P5EIS9 A0A182WJB0 A0A0P5KD98 A0A0P5FB89 A0A164MVI1 A0A0J9U9T9 A0A0P5X007 A0A0P4WEH3
PDB
5WM0
E-value=2.74328e-53,
Score=527
Ontologies
GO
Topology
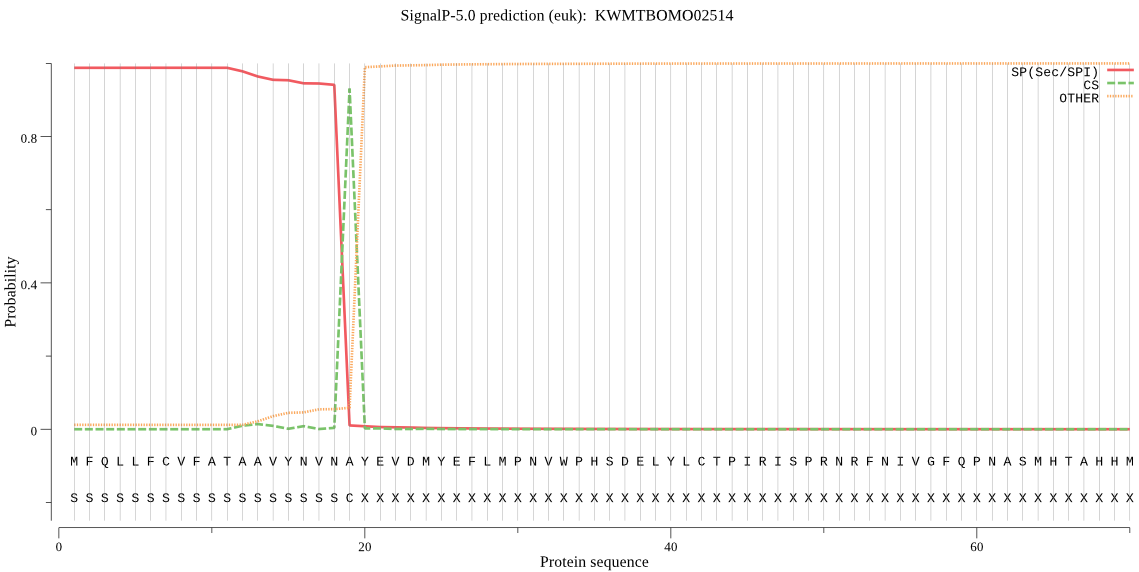
SignalP
Position: 1 - 19,
Likelihood: 0.987420
Length:
336
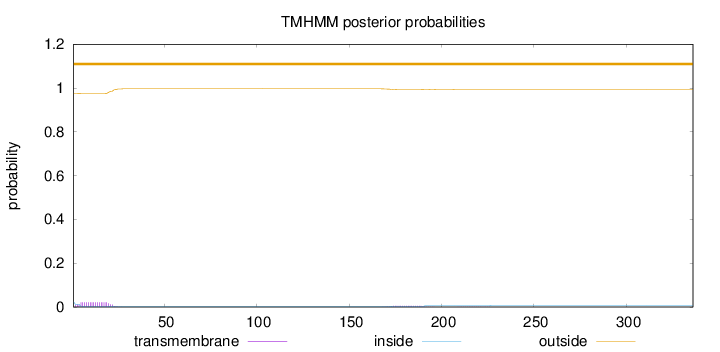
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.51993
Exp number, first 60 AAs:
0.41326
Total prob of N-in:
0.02400
outside
1 - 336
Population Genetic Test Statistics
Pi
319.626261
Theta
189.153383
Tajima's D
-0.994807
CLR
12.636218
CSRT
0.137293135343233
Interpretation
Uncertain
