Gene
KWMTBOMO02511
Pre Gene Modal
BGIBMGA002700
Annotation
PREDICTED:_tyrosine-protein_kinase_Drl-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.007 PlasmaMembrane Reliability : 1.438
Sequence
CDS
ATGGGAGGCTTAGACGCTGAGTTGTACTACGTTCGAGAAGGAGTCGTGAACACTTACGCTACGAGTTTCATTGTGCCCGTTCCGGCACATATCGCCGACTTGGAATTTATGTGGCAAGCCCTTGGACGAACACCGCTTCCGTACGTGATGGCTATAGAGTATGAGGCTAGAGGCGCCATGTTGCCGCCACAGGTCAATATCTCTGAACGGGGATATGTGCCCACAACTCTACAGACTTTTCGGATTCGTTTGCCGTGTACCGGGACGAGAAGTGCAGAAAACCTAGTCACAATTCAACTGAACATATCGGTTCCCGACCGCGCCCACAACGACATCCGATTGACCTTCAAAAGGAACAAGATCTGCTTAAAAGGCTTGGCGACAATAGTGACCCACAATGAGTCGGCACGTTTAGCCGGTGACGCTTCCCGTGGTCCGAGCGGAGTTCTATTCGCGGCGGGGGGCTGTGCGGCCGCAGCATTAGCGCTGCTCGCTGCCGCCGCCGCAGCCGGAACCCTGTACAAGCGGGGCAGCAAGCAAAGGCACCACGACTCGATACACAGTACGTCGTACACCACAGCGGCCTACGGGAGTCATCAAAACGTTCTCCTTAGATTGGATACGCTTGGTCGCCCGCCAAGTTCGACTGGATCCTACGCCACGATAGCTAGTTTACACAAATTTCCGTTTGGGAGTAGAAGATCACCATCGCCGTACGCAACAACGCAGATAGGTGAACATGTGTATGCGAAGCCGGATTCTCGCGTTTCGTACTACGCTGCGTCCAATCTAACTCACGTATCTCAGCAAACCGTTACAAAAGACCGCGTCACTGATCCAGCAGAACAACTTCGTGTTCTTACTACTTCTCGTTCGAACATACGCTTGGAGATTCTCATCCACGAAGGTACTTTCGGAAGAATCTATAAAGGGGTATTCAAAAAAGGCGATGTTTACGAAGAAGTTATGGTGAAGACTGTTTCAGATGCAGCGTCAGCTATGCAAGCTGCTCTACTTGTGGCAGAGGGATTAAGGTTATACGGACTTGTTCATGCAAACGTCCTAACACCAATGGCTGCATGTGCTGAAGAAGCGAGGAAACCACTTTTGATTTATTGCTGTCCTTCGCATTCTTCTAATCTAAAAAGGTTTTTGTCATCATGCCGTTTGGGTCAGCAGTCAGCTCCAGCAACTAGAGAACTGGTAGATTTAGGAGCACAGGTTGCTTGTGGATTATCTTATCTACATGTTCAACGATTTGTTCACGCTGATGTCGCTGCCAGAAATTGTGTGGTGGACGAAAAATTAAGACTGAAAGTAACGGATAATGGTTTAGCTAGAGACCTATTTCCTGATGATTACCATTGTCTTGGAGATAACGAGAATAGGCCTGTTAAATGGATGGCTCCGGAAACGTTACTTCAAAATCAATATTCTACTGCTTCGGATGTATGGTCGTTAGGAATAACCTTATGGGAACTGGCAACTCTGGGTTCCTCTCCATTGGCTGAACTTGATGCTGCTGACGTTGGAGCATTTTTAAACGGAGGCTATAGACCATCACAACCTCACAACTGTCCGGATGAATTGTATGGACTTATGTCTTGGTGTTGGACGACTCCGCCTTCAACACGACCAACTGCTCCTCAACTGCTAGCTGCACTTCACGACTTCAGGCAAGCGTTAAGCACTTATATATAA
Protein
MGGLDAELYYVREGVVNTYATSFIVPVPAHIADLEFMWQALGRTPLPYVMAIEYEARGAMLPPQVNISERGYVPTTLQTFRIRLPCTGTRSAENLVTIQLNISVPDRAHNDIRLTFKRNKICLKGLATIVTHNESARLAGDASRGPSGVLFAAGGCAAAALALLAAAAAAGTLYKRGSKQRHHDSIHSTSYTTAAYGSHQNVLLRLDTLGRPPSSTGSYATIASLHKFPFGSRRSPSPYATTQIGEHVYAKPDSRVSYYAASNLTHVSQQTVTKDRVTDPAEQLRVLTTSRSNIRLEILIHEGTFGRIYKGVFKKGDVYEEVMVKTVSDAASAMQAALLVAEGLRLYGLVHANVLTPMAACAEEARKPLLIYCCPSHSSNLKRFLSSCRLGQQSAPATRELVDLGAQVACGLSYLHVQRFVHADVAARNCVVDEKLRLKVTDNGLARDLFPDDYHCLGDNENRPVKWMAPETLLQNQYSTASDVWSLGITLWELATLGSSPLAELDAADVGAFLNGGYRPSQPHNCPDELYGLMSWCWTTPPSTRPTAPQLLAALHDFRQALSTYI
Summary
Uniprot
A0A194PEL2
A0A194QWR8
A0A212FKY8
A0A067R304
A0A1B6LUN1
A0A1B6FG07
+ More
A0A1B6CAR3 A0A158NVL7 A0A3L8D3K7 A0A0L0CGC1 A0A195BT17 A0A182UV33 W8BJC1 A0A084VEV1 A0A1Y9IVL6 A0A1I8MWH2 A0A151K0U7 A0A1I8PE35 W5J1L4 A0A1I8PDT5 A0A182QRR4 K7IWM0 A0A0M4E829 A0A151X5B5 A0A0J7L639 A0A1B6LZC5 E2A9X2 A0A2Y9D0T9 A0A182YQ99 A0A2J7QF99 A0A088ABL7 A0A0N0U4I8 A0A0T6AUW0 E0W415 A0A154P1L5 A0A1W4Y4F0 A0A3B1J6M4 A0A087UTU6 A0A088AHA8 A0A2A3E8A5 A0A0C9RJZ1 A0A154P277 A0A2U9C6T5 A0A0F8ARZ1 A0A0F8AX68 A0A3B3C4G7 A0A3B4VHJ8 A0A3Q0J0S9 A0A1Y1LXG5 A0A158NVL2 A0A195BUU5 E2ABN1 F4WGZ4 A0A3L8D3R9 A0A3P8TU24 A0A336M573 E2BV04 A0A2I4CX38 A0A3P8TY81 A0A151K1G5 A0A3P9BQ41 A0A3P8R818 A0A3P9KMZ2 A0A3B4GZH0 A0A3B5AQQ2 A0A3B3YX77 H2UZL4 A0A151X5B3 A0A087XDK3 A0A3B3UYD4 A0A3P9PLS9 A0A3P8TTZ0 A0A3B4GTR8 A0A3P8R897 A0A3P9HEC2 H2UZL6 I3KT49 A0A195E3X7 F4WGY6 A0A315URH0 A0A3B3XMI7 A0A3B3W1G8 A0A1A8PGM1 A0A1A8EC46 A0A2I4CX49 A0A1A8MFZ8 A0A1A8RDD3 A0A1A8CYD8 A0A1A8K039 A0A1A8B4Y6 H2L616 H2L619 A0A3P9HEN9 A0A3P9KMW0 A0A1A7Y1Q7 A0A3P9HE35 A0A3P9M4A1 A0A1A8ESS4
A0A1B6CAR3 A0A158NVL7 A0A3L8D3K7 A0A0L0CGC1 A0A195BT17 A0A182UV33 W8BJC1 A0A084VEV1 A0A1Y9IVL6 A0A1I8MWH2 A0A151K0U7 A0A1I8PE35 W5J1L4 A0A1I8PDT5 A0A182QRR4 K7IWM0 A0A0M4E829 A0A151X5B5 A0A0J7L639 A0A1B6LZC5 E2A9X2 A0A2Y9D0T9 A0A182YQ99 A0A2J7QF99 A0A088ABL7 A0A0N0U4I8 A0A0T6AUW0 E0W415 A0A154P1L5 A0A1W4Y4F0 A0A3B1J6M4 A0A087UTU6 A0A088AHA8 A0A2A3E8A5 A0A0C9RJZ1 A0A154P277 A0A2U9C6T5 A0A0F8ARZ1 A0A0F8AX68 A0A3B3C4G7 A0A3B4VHJ8 A0A3Q0J0S9 A0A1Y1LXG5 A0A158NVL2 A0A195BUU5 E2ABN1 F4WGZ4 A0A3L8D3R9 A0A3P8TU24 A0A336M573 E2BV04 A0A2I4CX38 A0A3P8TY81 A0A151K1G5 A0A3P9BQ41 A0A3P8R818 A0A3P9KMZ2 A0A3B4GZH0 A0A3B5AQQ2 A0A3B3YX77 H2UZL4 A0A151X5B3 A0A087XDK3 A0A3B3UYD4 A0A3P9PLS9 A0A3P8TTZ0 A0A3B4GTR8 A0A3P8R897 A0A3P9HEC2 H2UZL6 I3KT49 A0A195E3X7 F4WGY6 A0A315URH0 A0A3B3XMI7 A0A3B3W1G8 A0A1A8PGM1 A0A1A8EC46 A0A2I4CX49 A0A1A8MFZ8 A0A1A8RDD3 A0A1A8CYD8 A0A1A8K039 A0A1A8B4Y6 H2L616 H2L619 A0A3P9HEN9 A0A3P9KMW0 A0A1A7Y1Q7 A0A3P9HE35 A0A3P9M4A1 A0A1A8ESS4
Pubmed
EMBL
KQ459606
KPI91796.1
KQ461103
KPJ09405.1
AGBW02007973
OWR54360.1
+ More
KK852945 KDR13445.1 GEBQ01012591 JAT27386.1 GECZ01020705 JAS49064.1 GEDC01026933 JAS10365.1 ADTU01027256 ADTU01027257 ADTU01027258 ADTU01027259 ADTU01027260 ADTU01027261 ADTU01027262 ADTU01027263 ADTU01027264 QOIP01000014 RLU14794.1 JRES01000505 KNC30539.1 KQ976409 KYM90974.1 GAMC01007713 JAB98842.1 ATLV01012316 ATLV01012317 ATLV01012318 ATLV01012319 KE524780 KFB36495.1 KQ981248 KYN44237.1 ADMH02002166 ETN58092.1 AXCN02000882 CP012524 ALC40796.1 KQ982515 KYQ55586.1 LBMM01000546 KMQ98086.1 GEBQ01010993 JAT28984.1 GL437987 EFN69740.1 APCN01002242 APCN01002243 APCN01002244 APCN01002245 APCN01002246 NEVH01015301 PNF27271.1 KQ435815 KOX72651.1 LJIG01022756 KRT78888.1 DS235886 EEB20371.1 KQ434786 KZC05234.1 KK121583 KFM80785.1 KZ288347 PBC27512.1 GBYB01008515 JAG78282.1 KZC05230.1 CP026254 AWP11436.1 KQ041311 KKF28487.1 KQ042370 KKF15508.1 GEZM01049965 JAV75727.1 ADTU01027236 ADTU01027237 ADTU01027238 ADTU01027239 KYM90966.1 GL438334 EFN69133.1 GL888148 EGI66473.1 RLU15040.1 UFQT01000549 SSX25160.1 GL450776 EFN80484.1 KYN44243.1 KYQ55595.1 AYCK01014138 AERX01011523 AERX01011524 AERX01011525 AERX01011526 AERX01011527 KQ979701 KYN19617.1 EGI66465.1 NHOQ01002838 PWA14385.1 HAEG01007797 SBR80187.1 HAEA01014696 SBQ43176.1 HAEF01014140 SBR55299.1 HAEH01015728 HAEI01008220 SBS03393.1 HADZ01020117 SBP84058.1 HAED01022555 HAEE01005571 SBR25591.1 HADY01023834 HAEJ01016942 SBP62319.1 HADW01018443 HADX01001693 SBP23925.1 HAEB01003668 HAEC01016523 SBQ50195.1
KK852945 KDR13445.1 GEBQ01012591 JAT27386.1 GECZ01020705 JAS49064.1 GEDC01026933 JAS10365.1 ADTU01027256 ADTU01027257 ADTU01027258 ADTU01027259 ADTU01027260 ADTU01027261 ADTU01027262 ADTU01027263 ADTU01027264 QOIP01000014 RLU14794.1 JRES01000505 KNC30539.1 KQ976409 KYM90974.1 GAMC01007713 JAB98842.1 ATLV01012316 ATLV01012317 ATLV01012318 ATLV01012319 KE524780 KFB36495.1 KQ981248 KYN44237.1 ADMH02002166 ETN58092.1 AXCN02000882 CP012524 ALC40796.1 KQ982515 KYQ55586.1 LBMM01000546 KMQ98086.1 GEBQ01010993 JAT28984.1 GL437987 EFN69740.1 APCN01002242 APCN01002243 APCN01002244 APCN01002245 APCN01002246 NEVH01015301 PNF27271.1 KQ435815 KOX72651.1 LJIG01022756 KRT78888.1 DS235886 EEB20371.1 KQ434786 KZC05234.1 KK121583 KFM80785.1 KZ288347 PBC27512.1 GBYB01008515 JAG78282.1 KZC05230.1 CP026254 AWP11436.1 KQ041311 KKF28487.1 KQ042370 KKF15508.1 GEZM01049965 JAV75727.1 ADTU01027236 ADTU01027237 ADTU01027238 ADTU01027239 KYM90966.1 GL438334 EFN69133.1 GL888148 EGI66473.1 RLU15040.1 UFQT01000549 SSX25160.1 GL450776 EFN80484.1 KYN44243.1 KYQ55595.1 AYCK01014138 AERX01011523 AERX01011524 AERX01011525 AERX01011526 AERX01011527 KQ979701 KYN19617.1 EGI66465.1 NHOQ01002838 PWA14385.1 HAEG01007797 SBR80187.1 HAEA01014696 SBQ43176.1 HAEF01014140 SBR55299.1 HAEH01015728 HAEI01008220 SBS03393.1 HADZ01020117 SBP84058.1 HAED01022555 HAEE01005571 SBR25591.1 HADY01023834 HAEJ01016942 SBP62319.1 HADW01018443 HADX01001693 SBP23925.1 HAEB01003668 HAEC01016523 SBQ50195.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000027135
UP000005205
UP000279307
+ More
UP000037069 UP000078540 UP000075903 UP000030765 UP000075920 UP000095301 UP000078541 UP000095300 UP000000673 UP000075886 UP000002358 UP000092553 UP000075809 UP000036403 UP000000311 UP000075840 UP000076408 UP000235965 UP000005203 UP000053105 UP000009046 UP000076502 UP000192224 UP000018467 UP000054359 UP000242457 UP000246464 UP000261560 UP000261420 UP000079169 UP000007755 UP000265080 UP000008237 UP000192220 UP000265160 UP000265100 UP000265180 UP000261460 UP000261400 UP000261480 UP000005226 UP000028760 UP000261500 UP000242638 UP000265200 UP000005207 UP000078492 UP000001038
UP000037069 UP000078540 UP000075903 UP000030765 UP000075920 UP000095301 UP000078541 UP000095300 UP000000673 UP000075886 UP000002358 UP000092553 UP000075809 UP000036403 UP000000311 UP000075840 UP000076408 UP000235965 UP000005203 UP000053105 UP000009046 UP000076502 UP000192224 UP000018467 UP000054359 UP000242457 UP000246464 UP000261560 UP000261420 UP000079169 UP000007755 UP000265080 UP000008237 UP000192220 UP000265160 UP000265100 UP000265180 UP000261460 UP000261400 UP000261480 UP000005226 UP000028760 UP000261500 UP000242638 UP000265200 UP000005207 UP000078492 UP000001038
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
ProteinModelPortal
A0A194PEL2
A0A194QWR8
A0A212FKY8
A0A067R304
A0A1B6LUN1
A0A1B6FG07
+ More
A0A1B6CAR3 A0A158NVL7 A0A3L8D3K7 A0A0L0CGC1 A0A195BT17 A0A182UV33 W8BJC1 A0A084VEV1 A0A1Y9IVL6 A0A1I8MWH2 A0A151K0U7 A0A1I8PE35 W5J1L4 A0A1I8PDT5 A0A182QRR4 K7IWM0 A0A0M4E829 A0A151X5B5 A0A0J7L639 A0A1B6LZC5 E2A9X2 A0A2Y9D0T9 A0A182YQ99 A0A2J7QF99 A0A088ABL7 A0A0N0U4I8 A0A0T6AUW0 E0W415 A0A154P1L5 A0A1W4Y4F0 A0A3B1J6M4 A0A087UTU6 A0A088AHA8 A0A2A3E8A5 A0A0C9RJZ1 A0A154P277 A0A2U9C6T5 A0A0F8ARZ1 A0A0F8AX68 A0A3B3C4G7 A0A3B4VHJ8 A0A3Q0J0S9 A0A1Y1LXG5 A0A158NVL2 A0A195BUU5 E2ABN1 F4WGZ4 A0A3L8D3R9 A0A3P8TU24 A0A336M573 E2BV04 A0A2I4CX38 A0A3P8TY81 A0A151K1G5 A0A3P9BQ41 A0A3P8R818 A0A3P9KMZ2 A0A3B4GZH0 A0A3B5AQQ2 A0A3B3YX77 H2UZL4 A0A151X5B3 A0A087XDK3 A0A3B3UYD4 A0A3P9PLS9 A0A3P8TTZ0 A0A3B4GTR8 A0A3P8R897 A0A3P9HEC2 H2UZL6 I3KT49 A0A195E3X7 F4WGY6 A0A315URH0 A0A3B3XMI7 A0A3B3W1G8 A0A1A8PGM1 A0A1A8EC46 A0A2I4CX49 A0A1A8MFZ8 A0A1A8RDD3 A0A1A8CYD8 A0A1A8K039 A0A1A8B4Y6 H2L616 H2L619 A0A3P9HEN9 A0A3P9KMW0 A0A1A7Y1Q7 A0A3P9HE35 A0A3P9M4A1 A0A1A8ESS4
A0A1B6CAR3 A0A158NVL7 A0A3L8D3K7 A0A0L0CGC1 A0A195BT17 A0A182UV33 W8BJC1 A0A084VEV1 A0A1Y9IVL6 A0A1I8MWH2 A0A151K0U7 A0A1I8PE35 W5J1L4 A0A1I8PDT5 A0A182QRR4 K7IWM0 A0A0M4E829 A0A151X5B5 A0A0J7L639 A0A1B6LZC5 E2A9X2 A0A2Y9D0T9 A0A182YQ99 A0A2J7QF99 A0A088ABL7 A0A0N0U4I8 A0A0T6AUW0 E0W415 A0A154P1L5 A0A1W4Y4F0 A0A3B1J6M4 A0A087UTU6 A0A088AHA8 A0A2A3E8A5 A0A0C9RJZ1 A0A154P277 A0A2U9C6T5 A0A0F8ARZ1 A0A0F8AX68 A0A3B3C4G7 A0A3B4VHJ8 A0A3Q0J0S9 A0A1Y1LXG5 A0A158NVL2 A0A195BUU5 E2ABN1 F4WGZ4 A0A3L8D3R9 A0A3P8TU24 A0A336M573 E2BV04 A0A2I4CX38 A0A3P8TY81 A0A151K1G5 A0A3P9BQ41 A0A3P8R818 A0A3P9KMZ2 A0A3B4GZH0 A0A3B5AQQ2 A0A3B3YX77 H2UZL4 A0A151X5B3 A0A087XDK3 A0A3B3UYD4 A0A3P9PLS9 A0A3P8TTZ0 A0A3B4GTR8 A0A3P8R897 A0A3P9HEC2 H2UZL6 I3KT49 A0A195E3X7 F4WGY6 A0A315URH0 A0A3B3XMI7 A0A3B3W1G8 A0A1A8PGM1 A0A1A8EC46 A0A2I4CX49 A0A1A8MFZ8 A0A1A8RDD3 A0A1A8CYD8 A0A1A8K039 A0A1A8B4Y6 H2L616 H2L619 A0A3P9HEN9 A0A3P9KMW0 A0A1A7Y1Q7 A0A3P9HE35 A0A3P9M4A1 A0A1A8ESS4
PDB
3C1X
E-value=4.18914e-43,
Score=441
Ontologies
GO
Topology
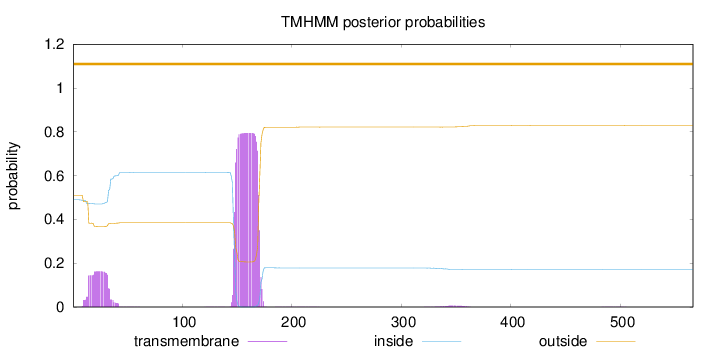
Length:
566
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
21.73133
Exp number, first 60 AAs:
3.36907
Total prob of N-in:
0.48950
outside
1 - 566
Population Genetic Test Statistics
Pi
237.504237
Theta
196.082765
Tajima's D
0.239327
CLR
0.723653
CSRT
0.437878106094695
Interpretation
Uncertain
