Gene
KWMTBOMO02509
Pre Gene Modal
BGIBMGA002702
Annotation
PREDICTED:_ken_and_barbie_protein_isoform_X1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.485
Sequence
CDS
ATGATGGGCGAGGGCCTGCTGACCTTGACGTACGGCAAGCACCACGCCTGCATCCTGAACGAGGTGGGCGCGGCGTGGCGCGGCGCCAGGTACTCCGACCTGGTGCTGGTCTGCGACGACGCCGTGCCCCTCGCGGCGCACAGAATCGTCATGGCCGCTGCTAGTCCTCTCATCAGGAAAATCCTCGACGATACACCATCAGTCGAGAATCCGGTCACGATTCACATGTCGGGCATCAACAGCACTCTCATGAGGCACCTACTTGTTTTTCTCTACAGCGGCCAAGCTTACATCGAGTCCGGCGAAATAGATGATATGCAAGAGCTGTTCGAATTATTAGAAATTAAATCGGACGTGTGGAAATCAACGAAAGAACGTCAGCAAGCCGAAGAAAGGAATAGGTCGTGTGAAAGAAAAGCAAAAAACCTCAAAACAGATATAAACAGCAATGAAAGCAGTAAACACGATGCGCCGTCTGGATCGTCACATTCAGAAAGTGAACGGGTCACACCGAGCGCAGGTTACAGTAAAGACAAAGACAAAGGAGACGCAGAATCGCTAAGTATAAAAAAAGAAAGTATGTCAACTAGCAGTAATGATGAAAGGGAAGATGAAGACGGAGAAAACAAAGAATGTGGCAGATTGACGGCACGGCGGCGGAGTTCCTCTAATCCTGTCAATCTATCTCTCGCACGAAATTCAGTAAACACAGGCAGTGAACATGACGACTCACAAGACGTAGATGTAGAAACAGTAGCAGCAAAGAATATATCTCGCCGGAGATCAACTTCATCAAGTCAAGACGATCAACCACAAGAAACGGTTTACAGGCGGCAGTTGCTCAATGACCGGATAGGTCGGGGGATTGAGAGAGCAAAAAGAAAATCACATTATTTGGAACCACCCGAATTGGATTTACGTACTGTTAAATTAGAAGATTACGCACACCTTAAACCGCCCGATCAAGATCTTATGTCATTGGTACAGACGTCTCCGGAAAATTATGTAGTCACACCACATCGGAAAAGAAGACCCGGCTTCCATAATTCACCTTCGCAAAATCCCCCGTTTGTTTCATTTCCACCAAGCTACTTAGAAGAAATGGCTCACCTACGGTATACTCAAGTACCGGCTACGGCTGCGGCTGTAGCGGCTAGTCGTCTAGGAGCATTACATTCACTAAGCGCATCAGCCCCTCCATATTTACCAGATCGAAGCCTAACACCCCCAACAGCCACACATGAAGAAGCGTTAAAATATAGACCGCCCAGCGCCGGATCGTGGGGACCATGGCTCTGTCAACCACAAATGGGAGGCGATGAAGCACCAGCGACGGAGCACGACCAAGGATCAAGTAAACAGGCACCAGTAAGAGAATATCGTTGTGAATATTGCGGGAAACAATTCGGCATGTCTTGGAATCTCAAAACCCATCTACGTGTACACACCGGAGAGAAACCGTTCGCTTGTCGTCTCTGCGTTGCTATGTTCAAACAAAAGGCACATTTACTGAAACATTTGTGTTCGGTACATCGTAACGTGATATCGTCAAGTGAAAACGACGGGAGAACTAACACGCCCGGACGGTTTAACTGTTGCTTCTGTCAACTTACCTTTGAAGCATTACCGGAGCTCATCAGACATTTGTCGGGACCACATAATAGTTTGCTTCTCAGCAAAAATTTGCACGAATGA
Protein
MMGEGLLTLTYGKHHACILNEVGAAWRGARYSDLVLVCDDAVPLAAHRIVMAAASPLIRKILDDTPSVENPVTIHMSGINSTLMRHLLVFLYSGQAYIESGEIDDMQELFELLEIKSDVWKSTKERQQAEERNRSCERKAKNLKTDINSNESSKHDAPSGSSHSESERVTPSAGYSKDKDKGDAESLSIKKESMSTSSNDEREDEDGENKECGRLTARRRSSSNPVNLSLARNSVNTGSEHDDSQDVDVETVAAKNISRRRSTSSSQDDQPQETVYRRQLLNDRIGRGIERAKRKSHYLEPPELDLRTVKLEDYAHLKPPDQDLMSLVQTSPENYVVTPHRKRRPGFHNSPSQNPPFVSFPPSYLEEMAHLRYTQVPATAAAVAASRLGALHSLSASAPPYLPDRSLTPPTATHEEALKYRPPSAGSWGPWLCQPQMGGDEAPATEHDQGSSKQAPVREYRCEYCGKQFGMSWNLKTHLRVHTGEKPFACRLCVAMFKQKAHLLKHLCSVHRNVISSSENDGRTNTPGRFNCCFCQLTFEALPELIRHLSGPHNSLLLSKNLHE
Summary
Uniprot
H9IZL7
F6ME18
A0A2A4JV34
A0A194PL12
A0A194R0N8
A0A212FG95
+ More
A0A0L7LH87 D7EIK2 N6TL45 A0A1Y1KRQ0 A0A0T6AUX3 N6U167 A0A1W4WRD8 A0A1B6CPX5 A0A067R5M6 A0A2J7Q100 A0A1B6GF48 A0A1B6LUC6 A0A0L7QP34 A0A0N0BF70 A0A088AHA5 A0A195CB08 F4WGY9 A0A151K0K1 E2BV01 A0A158NVL6 A0A023EYU8 A0A0C9QL96 A0A336LSP0 E9INZ7 A0A1B6JDK3 A0A1L8DIH3 E2ABM9 A0A3L8D2R4 A0A0A9XHE1 A0A146LLZ5 A0A0J7KX01 A0A154P047 A0A3B0JD63 B4LNX8 E0VT69 Q17I27 A0A0L0CNH1 A0A034W3J5 A0A0K8UP35 A0A0K8VTT3 A0A034W0Y5 A0A1A9WAE1 A0A1A9UYE7 A0A1A9YJN1 A0A1A9ZRD0 A0A182LI65 A0A0C9RBJ6 A0A0C9RSP8 A0A0K8SBC4 A0A195BUV0 T1HJM4 A0A1B0ARM3 A0A195E3G8
A0A0L7LH87 D7EIK2 N6TL45 A0A1Y1KRQ0 A0A0T6AUX3 N6U167 A0A1W4WRD8 A0A1B6CPX5 A0A067R5M6 A0A2J7Q100 A0A1B6GF48 A0A1B6LUC6 A0A0L7QP34 A0A0N0BF70 A0A088AHA5 A0A195CB08 F4WGY9 A0A151K0K1 E2BV01 A0A158NVL6 A0A023EYU8 A0A0C9QL96 A0A336LSP0 E9INZ7 A0A1B6JDK3 A0A1L8DIH3 E2ABM9 A0A3L8D2R4 A0A0A9XHE1 A0A146LLZ5 A0A0J7KX01 A0A154P047 A0A3B0JD63 B4LNX8 E0VT69 Q17I27 A0A0L0CNH1 A0A034W3J5 A0A0K8UP35 A0A0K8VTT3 A0A034W0Y5 A0A1A9WAE1 A0A1A9UYE7 A0A1A9YJN1 A0A1A9ZRD0 A0A182LI65 A0A0C9RBJ6 A0A0C9RSP8 A0A0K8SBC4 A0A195BUV0 T1HJM4 A0A1B0ARM3 A0A195E3G8
Pubmed
EMBL
BABH01014463
JF795585
AEG19520.1
NWSH01000617
PCG75270.1
KQ459606
+ More
KPI91795.1 KQ461103 KPJ09406.1 AGBW02008706 OWR52733.1 JTDY01001081 KOB74933.1 KQ971307 EFA12027.1 APGK01020372 KB740193 KB631602 ENN81159.1 ERL84556.1 GEZM01075750 JAV63984.1 LJIG01022756 KRT78889.1 APGK01047308 KB741077 ENN74361.1 GEDC01021756 JAS15542.1 KK852945 KDR13444.1 NEVH01019960 PNF22246.1 GECZ01008836 JAS60933.1 GEBQ01012690 JAT27287.1 KQ414836 KOC60380.1 KQ435815 KOX72648.1 KQ978072 KYM97293.1 GL888148 EGI66468.1 KQ981248 KYN44240.1 GL450776 EFN80481.1 ADTU01027243 ADTU01027244 GBBI01004344 JAC14368.1 GBYB01001367 JAG71134.1 UFQT01000092 SSX19599.1 GL764397 EFZ17724.1 GECU01010396 JAS97310.1 GFDF01007805 JAV06279.1 GL438334 EFN69131.1 QOIP01000014 RLU14795.1 GBHO01023497 GBRD01005451 JAG20107.1 JAG60370.1 GDHC01009575 JAQ09054.1 LBMM01002434 KMQ94841.1 KQ434786 KZC05231.1 OUUW01000001 SPP73160.1 CH940648 EDW61147.1 DS235760 EEB16575.1 CH477243 EAT46332.1 JRES01000153 KNC33772.1 GAKP01010609 JAC48343.1 GDHF01024194 GDHF01006514 JAI28120.1 JAI45800.1 GDHF01010051 JAI42263.1 GAKP01010608 JAC48344.1 GBYB01010507 JAG80274.1 GBYB01010506 JAG80273.1 GBRD01015781 JAG50045.1 KQ976409 KYM90971.1 ACPB03007777 JXJN01002483 KQ979701 KYN19621.1
KPI91795.1 KQ461103 KPJ09406.1 AGBW02008706 OWR52733.1 JTDY01001081 KOB74933.1 KQ971307 EFA12027.1 APGK01020372 KB740193 KB631602 ENN81159.1 ERL84556.1 GEZM01075750 JAV63984.1 LJIG01022756 KRT78889.1 APGK01047308 KB741077 ENN74361.1 GEDC01021756 JAS15542.1 KK852945 KDR13444.1 NEVH01019960 PNF22246.1 GECZ01008836 JAS60933.1 GEBQ01012690 JAT27287.1 KQ414836 KOC60380.1 KQ435815 KOX72648.1 KQ978072 KYM97293.1 GL888148 EGI66468.1 KQ981248 KYN44240.1 GL450776 EFN80481.1 ADTU01027243 ADTU01027244 GBBI01004344 JAC14368.1 GBYB01001367 JAG71134.1 UFQT01000092 SSX19599.1 GL764397 EFZ17724.1 GECU01010396 JAS97310.1 GFDF01007805 JAV06279.1 GL438334 EFN69131.1 QOIP01000014 RLU14795.1 GBHO01023497 GBRD01005451 JAG20107.1 JAG60370.1 GDHC01009575 JAQ09054.1 LBMM01002434 KMQ94841.1 KQ434786 KZC05231.1 OUUW01000001 SPP73160.1 CH940648 EDW61147.1 DS235760 EEB16575.1 CH477243 EAT46332.1 JRES01000153 KNC33772.1 GAKP01010609 JAC48343.1 GDHF01024194 GDHF01006514 JAI28120.1 JAI45800.1 GDHF01010051 JAI42263.1 GAKP01010608 JAC48344.1 GBYB01010507 JAG80274.1 GBYB01010506 JAG80273.1 GBRD01015781 JAG50045.1 KQ976409 KYM90971.1 ACPB03007777 JXJN01002483 KQ979701 KYN19621.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000007266 UP000019118 UP000030742 UP000192223 UP000027135 UP000235965 UP000053825 UP000053105 UP000005203 UP000078542 UP000007755 UP000078541 UP000008237 UP000005205 UP000000311 UP000279307 UP000036403 UP000076502 UP000268350 UP000008792 UP000009046 UP000008820 UP000037069 UP000091820 UP000078200 UP000092443 UP000092445 UP000075882 UP000078540 UP000015103 UP000092460 UP000078492
UP000007266 UP000019118 UP000030742 UP000192223 UP000027135 UP000235965 UP000053825 UP000053105 UP000005203 UP000078542 UP000007755 UP000078541 UP000008237 UP000005205 UP000000311 UP000279307 UP000036403 UP000076502 UP000268350 UP000008792 UP000009046 UP000008820 UP000037069 UP000091820 UP000078200 UP000092443 UP000092445 UP000075882 UP000078540 UP000015103 UP000092460 UP000078492
Interpro
ProteinModelPortal
H9IZL7
F6ME18
A0A2A4JV34
A0A194PL12
A0A194R0N8
A0A212FG95
+ More
A0A0L7LH87 D7EIK2 N6TL45 A0A1Y1KRQ0 A0A0T6AUX3 N6U167 A0A1W4WRD8 A0A1B6CPX5 A0A067R5M6 A0A2J7Q100 A0A1B6GF48 A0A1B6LUC6 A0A0L7QP34 A0A0N0BF70 A0A088AHA5 A0A195CB08 F4WGY9 A0A151K0K1 E2BV01 A0A158NVL6 A0A023EYU8 A0A0C9QL96 A0A336LSP0 E9INZ7 A0A1B6JDK3 A0A1L8DIH3 E2ABM9 A0A3L8D2R4 A0A0A9XHE1 A0A146LLZ5 A0A0J7KX01 A0A154P047 A0A3B0JD63 B4LNX8 E0VT69 Q17I27 A0A0L0CNH1 A0A034W3J5 A0A0K8UP35 A0A0K8VTT3 A0A034W0Y5 A0A1A9WAE1 A0A1A9UYE7 A0A1A9YJN1 A0A1A9ZRD0 A0A182LI65 A0A0C9RBJ6 A0A0C9RSP8 A0A0K8SBC4 A0A195BUV0 T1HJM4 A0A1B0ARM3 A0A195E3G8
A0A0L7LH87 D7EIK2 N6TL45 A0A1Y1KRQ0 A0A0T6AUX3 N6U167 A0A1W4WRD8 A0A1B6CPX5 A0A067R5M6 A0A2J7Q100 A0A1B6GF48 A0A1B6LUC6 A0A0L7QP34 A0A0N0BF70 A0A088AHA5 A0A195CB08 F4WGY9 A0A151K0K1 E2BV01 A0A158NVL6 A0A023EYU8 A0A0C9QL96 A0A336LSP0 E9INZ7 A0A1B6JDK3 A0A1L8DIH3 E2ABM9 A0A3L8D2R4 A0A0A9XHE1 A0A146LLZ5 A0A0J7KX01 A0A154P047 A0A3B0JD63 B4LNX8 E0VT69 Q17I27 A0A0L0CNH1 A0A034W3J5 A0A0K8UP35 A0A0K8VTT3 A0A034W0Y5 A0A1A9WAE1 A0A1A9UYE7 A0A1A9YJN1 A0A1A9ZRD0 A0A182LI65 A0A0C9RBJ6 A0A0C9RSP8 A0A0K8SBC4 A0A195BUV0 T1HJM4 A0A1B0ARM3 A0A195E3G8
PDB
2M9A
E-value=5.23389e-08,
Score=139
Ontologies
GO
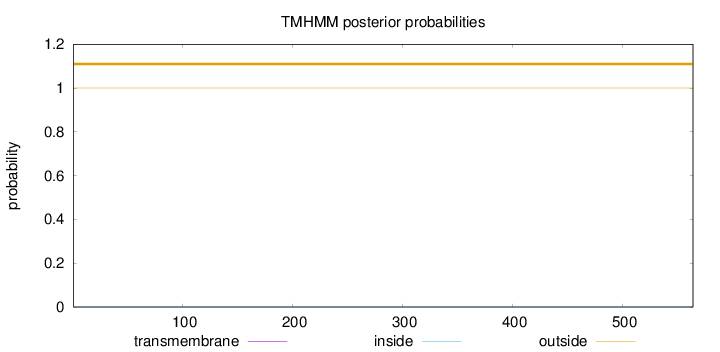
Topology
Length:
564
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00354000000000001
Exp number, first 60 AAs:
0.0014
Total prob of N-in:
0.00029
outside
1 - 564
Population Genetic Test Statistics
Pi
167.857935
Theta
188.107325
Tajima's D
-1.011681
CLR
1018.745256
CSRT
0.137343132843358
Interpretation
Uncertain
