Gene
KWMTBOMO02508 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002703
Annotation
32_kDa_apolipoprotein_precursor_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.884
Sequence
CDS
ATGGCGAATTTCAACATGAACCGGTTTTTGGGTACTTGGTATGAAGCCGAGCGGTTCTTCACCGTCAGCGAACTTGGCTCTAGATGCGTCACCACCAACTATGTGAGCACTCCTGAGGGAAGAATCATTGTATCCAATGAAATCGTCAATTCTTTAACGGGAATGAAACGCCTTATGGAAGGTAGCCTTCAGATGATAGGGCGCGAAGGCGAGGGACGCTTCATGATTAAGTACTCATCATTGCCACTTCCGTATGAGTCCGAGTTCTCAATCCTGGACACGGACTACGACAACTATGCGGTGATGTGGTCATGCAGCGGCATAGGGCCGGTTCACACTCGTAAGTTATCGATGAAAGTATTCATATAA
Protein
MANFNMNRFLGTWYEAERFFTVSELGSRCVTTNYVSTPEGRIIVSNEIVNSLTGMKRLMEGSLQMIGREGEGRFMIKYSSLPLPYESEFSILDTDYDNYAVMWSCSGIGPVHTRKLSMKVFI
Summary
Similarity
Belongs to the calycin superfamily. Lipocalin family.
Uniprot
H9IZL8
B9VTR5
A0A194QVB0
Q8T5Q9
A0A2A4JUV9
A0A2H1WZF6
+ More
A0A194PGG0 A0A212FG74 A0A2A4JUH0 A0A1D1ZII0 D6W7I9 A0A1W4WNZ9 E0VT70 J3JV57 A0A1B6CC82 A0A0K8TIR9 A0A0L7QPH9 A0A161MKB5 A0A2R7VY94 R4G7P7 A3EXQ4 A0A0T6BF28 A0A1Y1KWD0 A0A0J7NQV0 A0A154P066 A0A069DSJ7 F4WGZ0 A0A151X5N1 A0A195E378 A0A151K0M4 A0A195C8Y6 A0A3Q0J714 E2ABN0 E2BV02 A0A2A3E7M9 A0A232ERQ4 A0A088AHA6 E9INZ6 A0A0M8ZX05 A0A0C9R4U6 A0A2S2NLC9 A0A2H8TJV2 J9K7R3 A0A3L8D3H1 A0A026X1L2 A0A158NVL4 T1GZ20 A0A1J1IQQ4 B0X2C5 B0WRA7 A0A2P8XUJ1 Q170B5 A0A336LF49 A0A2J7QF90 A0A2P8XY88 A0A0T6AWB3 A0A182VUI2 A0A1W7R7Q0 A0A182RAY1 A0A182F4R4 A0A182YDP1 Q7PVG8 A0A067R6A5 A0A182L6G2 A0A182PG15 A0A182UZV9 A0A182X0R9 A0A182HGF6 W5J8E2 A0A182UD40 A0A151K0W3 A0A1S4H1E9 A0A151X5C5 A0A195E432 F4WGZ1 A0A195CAR7 A0A182Q6U5 A0A158NVL3 E2BV03 A0A182NU62 A0A195BU21 A0A084VWH5 A0A0J7KX35 A0A1B0ETQ6 A0A154P065 A0A182J3G0 H9IZL9 C4WT38 A0A3L8D4I0 A0A212EL40 A0A2A4JTA8 A0A2J7Q0Z2 A0A0M8ZWP7 A0A067QW30 A0A0C9R3T7 A0A0C9R3U4 K7IWM2 A0A232ERR7 A0A2A3E6W9 A0A088AHA7
A0A194PGG0 A0A212FG74 A0A2A4JUH0 A0A1D1ZII0 D6W7I9 A0A1W4WNZ9 E0VT70 J3JV57 A0A1B6CC82 A0A0K8TIR9 A0A0L7QPH9 A0A161MKB5 A0A2R7VY94 R4G7P7 A3EXQ4 A0A0T6BF28 A0A1Y1KWD0 A0A0J7NQV0 A0A154P066 A0A069DSJ7 F4WGZ0 A0A151X5N1 A0A195E378 A0A151K0M4 A0A195C8Y6 A0A3Q0J714 E2ABN0 E2BV02 A0A2A3E7M9 A0A232ERQ4 A0A088AHA6 E9INZ6 A0A0M8ZX05 A0A0C9R4U6 A0A2S2NLC9 A0A2H8TJV2 J9K7R3 A0A3L8D3H1 A0A026X1L2 A0A158NVL4 T1GZ20 A0A1J1IQQ4 B0X2C5 B0WRA7 A0A2P8XUJ1 Q170B5 A0A336LF49 A0A2J7QF90 A0A2P8XY88 A0A0T6AWB3 A0A182VUI2 A0A1W7R7Q0 A0A182RAY1 A0A182F4R4 A0A182YDP1 Q7PVG8 A0A067R6A5 A0A182L6G2 A0A182PG15 A0A182UZV9 A0A182X0R9 A0A182HGF6 W5J8E2 A0A182UD40 A0A151K0W3 A0A1S4H1E9 A0A151X5C5 A0A195E432 F4WGZ1 A0A195CAR7 A0A182Q6U5 A0A158NVL3 E2BV03 A0A182NU62 A0A195BU21 A0A084VWH5 A0A0J7KX35 A0A1B0ETQ6 A0A154P065 A0A182J3G0 H9IZL9 C4WT38 A0A3L8D4I0 A0A212EL40 A0A2A4JTA8 A0A2J7Q0Z2 A0A0M8ZWP7 A0A067QW30 A0A0C9R3T7 A0A0C9R3U4 K7IWM2 A0A232ERR7 A0A2A3E6W9 A0A088AHA7
Pubmed
EMBL
BABH01014467
FJ602775
ACM24341.1
KQ461103
KPJ09407.1
AF497850
+ More
AAM18117.2 NWSH01000617 PCG75273.1 ODYU01012209 SOQ58392.1 KQ459606 KPI91794.1 AGBW02008706 OWR52732.1 PCG75274.1 GDJX01001287 JAT66649.1 KQ971307 EFA11278.1 DS235760 EEB16576.1 APGK01020372 APGK01047308 BT127123 KB741077 KB740193 KB631602 AEE62085.1 ENN74359.1 ENN81161.1 ERL84558.1 GEDC01026257 JAS11041.1 GBRD01000505 JAG65316.1 KQ414836 KOC60381.1 GEMB01000449 JAS02676.1 KK854167 PTY12473.1 ACPB03007775 GAHY01002352 JAA75158.1 EF091972 ABN11964.1 LJIG01001167 KRT85773.1 GEZM01075750 JAV63985.1 LBMM01002434 KMQ94840.1 KQ434786 KZC05232.1 GBGD01002014 JAC86875.1 GL888148 EGI66469.1 KQ982515 KYQ55590.1 KQ979701 KYN19620.1 KQ981248 KYN44241.1 KQ978072 KYM97292.1 GL438334 EFN69132.1 GL450776 EFN80482.1 KZ288347 PBC27514.1 NNAY01002561 OXU21044.1 GL764397 EFZ17684.1 KQ435815 KOX72647.1 GBYB01011314 JAG81081.1 GGMR01005366 MBY17985.1 GFXV01002514 MBW14319.1 ABLF02011171 QOIP01000014 RLU14796.1 KK107061 EZA61279.1 ADTU01027240 ADTU01027241 ADTU01027242 CAQQ02388143 CVRI01000054 CRL00825.1 DS232286 EDS39148.1 DS232053 EDS33291.1 PYGN01001323 PSN35678.1 CH477477 EAT40277.1 UFQS01004093 UFQT01004093 SSX16169.1 SSX35495.1 NEVH01015301 PNF27250.1 PYGN01001171 PSN36978.1 LJIG01022756 KRT78887.1 GEHC01000520 JAV47125.1 AAAB01008984 EAA15071.4 KK852670 KDR18800.1 APCN01004224 ADMH02001962 ETN60231.1 KYN44242.1 KYQ55591.1 KYN19619.1 EGI66470.1 KYM97291.1 AXCN02000773 EFN80483.1 KQ976409 KYM90970.1 ATLV01017588 KE525174 KFB42319.1 KMQ94839.1 AJWK01010323 AJWK01010324 KZC05233.1 AK340452 BAH71058.1 RLU15041.1 AGBW02014147 OWR42187.1 PCG75275.1 PCG75276.1 NEVH01019960 PNF22244.1 KOX72645.1 LC382016 KK852945 BBE00746.1 KDR13443.1 GBYB01001481 JAG71248.1 GBYB01001486 JAG71253.1 OXU21045.1 PBC27513.1
AAM18117.2 NWSH01000617 PCG75273.1 ODYU01012209 SOQ58392.1 KQ459606 KPI91794.1 AGBW02008706 OWR52732.1 PCG75274.1 GDJX01001287 JAT66649.1 KQ971307 EFA11278.1 DS235760 EEB16576.1 APGK01020372 APGK01047308 BT127123 KB741077 KB740193 KB631602 AEE62085.1 ENN74359.1 ENN81161.1 ERL84558.1 GEDC01026257 JAS11041.1 GBRD01000505 JAG65316.1 KQ414836 KOC60381.1 GEMB01000449 JAS02676.1 KK854167 PTY12473.1 ACPB03007775 GAHY01002352 JAA75158.1 EF091972 ABN11964.1 LJIG01001167 KRT85773.1 GEZM01075750 JAV63985.1 LBMM01002434 KMQ94840.1 KQ434786 KZC05232.1 GBGD01002014 JAC86875.1 GL888148 EGI66469.1 KQ982515 KYQ55590.1 KQ979701 KYN19620.1 KQ981248 KYN44241.1 KQ978072 KYM97292.1 GL438334 EFN69132.1 GL450776 EFN80482.1 KZ288347 PBC27514.1 NNAY01002561 OXU21044.1 GL764397 EFZ17684.1 KQ435815 KOX72647.1 GBYB01011314 JAG81081.1 GGMR01005366 MBY17985.1 GFXV01002514 MBW14319.1 ABLF02011171 QOIP01000014 RLU14796.1 KK107061 EZA61279.1 ADTU01027240 ADTU01027241 ADTU01027242 CAQQ02388143 CVRI01000054 CRL00825.1 DS232286 EDS39148.1 DS232053 EDS33291.1 PYGN01001323 PSN35678.1 CH477477 EAT40277.1 UFQS01004093 UFQT01004093 SSX16169.1 SSX35495.1 NEVH01015301 PNF27250.1 PYGN01001171 PSN36978.1 LJIG01022756 KRT78887.1 GEHC01000520 JAV47125.1 AAAB01008984 EAA15071.4 KK852670 KDR18800.1 APCN01004224 ADMH02001962 ETN60231.1 KYN44242.1 KYQ55591.1 KYN19619.1 EGI66470.1 KYM97291.1 AXCN02000773 EFN80483.1 KQ976409 KYM90970.1 ATLV01017588 KE525174 KFB42319.1 KMQ94839.1 AJWK01010323 AJWK01010324 KZC05233.1 AK340452 BAH71058.1 RLU15041.1 AGBW02014147 OWR42187.1 PCG75275.1 PCG75276.1 NEVH01019960 PNF22244.1 KOX72645.1 LC382016 KK852945 BBE00746.1 KDR13443.1 GBYB01001481 JAG71248.1 GBYB01001486 JAG71253.1 OXU21045.1 PBC27513.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000007266
+ More
UP000192223 UP000009046 UP000019118 UP000030742 UP000053825 UP000015103 UP000036403 UP000076502 UP000007755 UP000075809 UP000078492 UP000078541 UP000078542 UP000079169 UP000000311 UP000008237 UP000242457 UP000215335 UP000005203 UP000053105 UP000007819 UP000279307 UP000053097 UP000005205 UP000015102 UP000183832 UP000002320 UP000245037 UP000008820 UP000235965 UP000075920 UP000075900 UP000069272 UP000076408 UP000007062 UP000027135 UP000075882 UP000075885 UP000075903 UP000076407 UP000075840 UP000000673 UP000075902 UP000075886 UP000075884 UP000078540 UP000030765 UP000092461 UP000075880 UP000002358
UP000192223 UP000009046 UP000019118 UP000030742 UP000053825 UP000015103 UP000036403 UP000076502 UP000007755 UP000075809 UP000078492 UP000078541 UP000078542 UP000079169 UP000000311 UP000008237 UP000242457 UP000215335 UP000005203 UP000053105 UP000007819 UP000279307 UP000053097 UP000005205 UP000015102 UP000183832 UP000002320 UP000245037 UP000008820 UP000235965 UP000075920 UP000075900 UP000069272 UP000076408 UP000007062 UP000027135 UP000075882 UP000075885 UP000075903 UP000076407 UP000075840 UP000000673 UP000075902 UP000075886 UP000075884 UP000078540 UP000030765 UP000092461 UP000075880 UP000002358
Interpro
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
H9IZL8
B9VTR5
A0A194QVB0
Q8T5Q9
A0A2A4JUV9
A0A2H1WZF6
+ More
A0A194PGG0 A0A212FG74 A0A2A4JUH0 A0A1D1ZII0 D6W7I9 A0A1W4WNZ9 E0VT70 J3JV57 A0A1B6CC82 A0A0K8TIR9 A0A0L7QPH9 A0A161MKB5 A0A2R7VY94 R4G7P7 A3EXQ4 A0A0T6BF28 A0A1Y1KWD0 A0A0J7NQV0 A0A154P066 A0A069DSJ7 F4WGZ0 A0A151X5N1 A0A195E378 A0A151K0M4 A0A195C8Y6 A0A3Q0J714 E2ABN0 E2BV02 A0A2A3E7M9 A0A232ERQ4 A0A088AHA6 E9INZ6 A0A0M8ZX05 A0A0C9R4U6 A0A2S2NLC9 A0A2H8TJV2 J9K7R3 A0A3L8D3H1 A0A026X1L2 A0A158NVL4 T1GZ20 A0A1J1IQQ4 B0X2C5 B0WRA7 A0A2P8XUJ1 Q170B5 A0A336LF49 A0A2J7QF90 A0A2P8XY88 A0A0T6AWB3 A0A182VUI2 A0A1W7R7Q0 A0A182RAY1 A0A182F4R4 A0A182YDP1 Q7PVG8 A0A067R6A5 A0A182L6G2 A0A182PG15 A0A182UZV9 A0A182X0R9 A0A182HGF6 W5J8E2 A0A182UD40 A0A151K0W3 A0A1S4H1E9 A0A151X5C5 A0A195E432 F4WGZ1 A0A195CAR7 A0A182Q6U5 A0A158NVL3 E2BV03 A0A182NU62 A0A195BU21 A0A084VWH5 A0A0J7KX35 A0A1B0ETQ6 A0A154P065 A0A182J3G0 H9IZL9 C4WT38 A0A3L8D4I0 A0A212EL40 A0A2A4JTA8 A0A2J7Q0Z2 A0A0M8ZWP7 A0A067QW30 A0A0C9R3T7 A0A0C9R3U4 K7IWM2 A0A232ERR7 A0A2A3E6W9 A0A088AHA7
A0A194PGG0 A0A212FG74 A0A2A4JUH0 A0A1D1ZII0 D6W7I9 A0A1W4WNZ9 E0VT70 J3JV57 A0A1B6CC82 A0A0K8TIR9 A0A0L7QPH9 A0A161MKB5 A0A2R7VY94 R4G7P7 A3EXQ4 A0A0T6BF28 A0A1Y1KWD0 A0A0J7NQV0 A0A154P066 A0A069DSJ7 F4WGZ0 A0A151X5N1 A0A195E378 A0A151K0M4 A0A195C8Y6 A0A3Q0J714 E2ABN0 E2BV02 A0A2A3E7M9 A0A232ERQ4 A0A088AHA6 E9INZ6 A0A0M8ZX05 A0A0C9R4U6 A0A2S2NLC9 A0A2H8TJV2 J9K7R3 A0A3L8D3H1 A0A026X1L2 A0A158NVL4 T1GZ20 A0A1J1IQQ4 B0X2C5 B0WRA7 A0A2P8XUJ1 Q170B5 A0A336LF49 A0A2J7QF90 A0A2P8XY88 A0A0T6AWB3 A0A182VUI2 A0A1W7R7Q0 A0A182RAY1 A0A182F4R4 A0A182YDP1 Q7PVG8 A0A067R6A5 A0A182L6G2 A0A182PG15 A0A182UZV9 A0A182X0R9 A0A182HGF6 W5J8E2 A0A182UD40 A0A151K0W3 A0A1S4H1E9 A0A151X5C5 A0A195E432 F4WGZ1 A0A195CAR7 A0A182Q6U5 A0A158NVL3 E2BV03 A0A182NU62 A0A195BU21 A0A084VWH5 A0A0J7KX35 A0A1B0ETQ6 A0A154P065 A0A182J3G0 H9IZL9 C4WT38 A0A3L8D4I0 A0A212EL40 A0A2A4JTA8 A0A2J7Q0Z2 A0A0M8ZWP7 A0A067QW30 A0A0C9R3T7 A0A0C9R3U4 K7IWM2 A0A232ERR7 A0A2A3E6W9 A0A088AHA7
PDB
2HZR
E-value=2.02766e-10,
Score=151
Ontologies
KEGG
GO
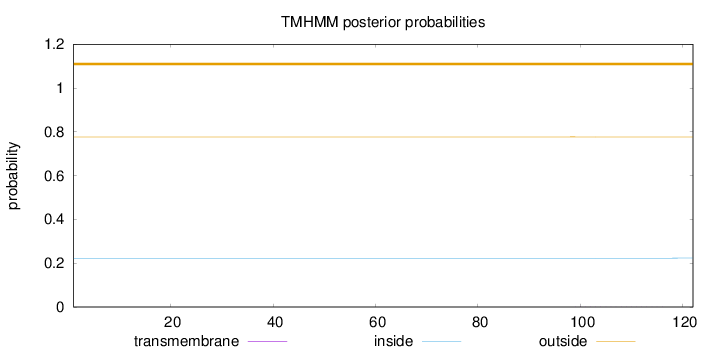
Topology
Length:
122
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00566
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.22314
outside
1 - 122
Population Genetic Test Statistics
Pi
208.063321
Theta
144.93097
Tajima's D
1.333272
CLR
0.997407
CSRT
0.749662516874156
Interpretation
Uncertain
