Gene
KWMTBOMO02495
Pre Gene Modal
BGIBMGA002711
Annotation
Rab-related_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.581 Mitochondrial Reliability : 1.677
Sequence
CDS
ATGTCGCCGAAACAAAAGCAACACGCCGGCAGTTCACACGCATCGGCTAGTGCTGAACGCCGGGAGCATTTATATAAGATCCTCGTCATCGGCGAATTGGGTACAGGGAAGACTTCCATAATCAAACGATATGTCCACCAGTTCTTCAGTCAACACTATAGAGCAACCATCGGCGTGGATTTCGCACTGAAAGTACTGAATTGGGACTCAAATACCATAATAAGATTGCAACTATGGGATATAGCAGGACAAGAGCGCTTCGGGAACATGACGCGTGTTTATTACAAGGAAGCAGTGGGCGCGTTTATAGTGTTCGACGTTTCCAGAGTGGCCACGTTCGACGCTGTCGTCAAGTGGAAGAATGACTTGGATGCGAAAGTTCAGTTGCCCGATGGATCGGCCATACCTTGTATACTGCTCGCTAATAAGTGCGATCAACAGAAGGAAGGTATAGTCAATAGTCCCGCCAAAATGGACGAGTACTGTCGCGAGAAAGGTTTCGCGGGCTGGTTCGAGACATCCGCCAAAGAGAACATCAATATTGAGGAGGCAGCTCGGTCATTAGTTAATAAGATTCTATTAAACGACAAGCTTCTTCAGAACAACGACACCAGGGACGGTGATAAATTCGTGATTGATCACAAAATAGCTAACGGTGACAGCAGAGATGGCGCGAACAAGGCTTGCGGTTGCTGA
Protein
MSPKQKQHAGSSHASASAERREHLYKILVIGELGTGKTSIIKRYVHQFFSQHYRATIGVDFALKVLNWDSNTIIRLQLWDIAGQERFGNMTRVYYKEAVGAFIVFDVSRVATFDAVVKWKNDLDAKVQLPDGSAIPCILLANKCDQQKEGIVNSPAKMDEYCREKGFAGWFETSAKENINIEEAARSLVNKILLNDKLLQNNDTRDGDKFVIDHKIANGDSRDGANKACGC
Summary
Uniprot
H9IZM6
Q2F5W5
F2VY73
A0A2A4JDA7
A0A2A4JCX7
A0A2A4JDB5
+ More
A0A194PEJ3 A0A194R0Q2 A0A212EPC9 A0A088AJT1 A0A0C9R9Y5 A0A0C9RW95 A0A067RKP3 E0VCG5 A0A182U6P0 Q7QJT8 A0A182S9B0 F4WDG0 A0A2A3EER7 A0A0M9A049 A0A182XZG9 A0A2J7Q8U2 E2A9Q7 A0A182JYQ1 A0A2J7Q8U7 A0A0L7QTZ9 E2BRQ4 A0A1B6FZT9 B0WZR8 A0A1Q3F6J3 A0A1B6GKK6 A0A1Q3F6U4 A0A3L8DKT4 A0A1B6IK06 A0A1Q3F6I0 A0A2M4BYC1 A0A1B6H7K9 A0A1B6LC41 T1E7S6 A0A195FRW0 A0A2J7Q8V1 B4QGT7 B4HSP2 A0A2M4BZF2 A0A1B6MK13 A1Z7S2 A0A151J7K8 A0A158NIR4 Q8IH37 A0A0J9R8X9 A1Z7S1 A0A0Q5WKM2 A1Z7S3 A0A0B4LEY0 Q7KY07 A0A023EK20 A0A0J9R8T1 A0A0J9TX72 T1E268 A0A1B6BYD4 A0A026WLZ2 A0A151X0T5 W8CBL4 A0A0R1DTY3 A0A0Q5WNC0 A0A1Y1KVJ8 A0A0J9R9W6 Q8T0G8 B3N7J6 Q86PD0 U5ERJ9 B4P3Z0 J9HYU4 Q174H5 A0A154PKM5 A0A1B6CA05 A0A0Q9XJX9 A0A0A1XRI0 A0A195C9U3 A0A1W4UY09 A0A3B0JJQ9 W8BKL3 A0A0B4UBW9 A0A1W4X2P4 B3MD31 N6W6G8 A0A0Q9XFQ4 A0A034VF73 A0A182R0W5 B4NNM5 A0A182X6G6 A0A182I3R9 A0A182VMA7 A0A1I8NYK8 V4B577 A0A1W4WT24 A0A2T7PEC0 T1PIC5 B4ME48 A0A182P9W2
A0A194PEJ3 A0A194R0Q2 A0A212EPC9 A0A088AJT1 A0A0C9R9Y5 A0A0C9RW95 A0A067RKP3 E0VCG5 A0A182U6P0 Q7QJT8 A0A182S9B0 F4WDG0 A0A2A3EER7 A0A0M9A049 A0A182XZG9 A0A2J7Q8U2 E2A9Q7 A0A182JYQ1 A0A2J7Q8U7 A0A0L7QTZ9 E2BRQ4 A0A1B6FZT9 B0WZR8 A0A1Q3F6J3 A0A1B6GKK6 A0A1Q3F6U4 A0A3L8DKT4 A0A1B6IK06 A0A1Q3F6I0 A0A2M4BYC1 A0A1B6H7K9 A0A1B6LC41 T1E7S6 A0A195FRW0 A0A2J7Q8V1 B4QGT7 B4HSP2 A0A2M4BZF2 A0A1B6MK13 A1Z7S2 A0A151J7K8 A0A158NIR4 Q8IH37 A0A0J9R8X9 A1Z7S1 A0A0Q5WKM2 A1Z7S3 A0A0B4LEY0 Q7KY07 A0A023EK20 A0A0J9R8T1 A0A0J9TX72 T1E268 A0A1B6BYD4 A0A026WLZ2 A0A151X0T5 W8CBL4 A0A0R1DTY3 A0A0Q5WNC0 A0A1Y1KVJ8 A0A0J9R9W6 Q8T0G8 B3N7J6 Q86PD0 U5ERJ9 B4P3Z0 J9HYU4 Q174H5 A0A154PKM5 A0A1B6CA05 A0A0Q9XJX9 A0A0A1XRI0 A0A195C9U3 A0A1W4UY09 A0A3B0JJQ9 W8BKL3 A0A0B4UBW9 A0A1W4X2P4 B3MD31 N6W6G8 A0A0Q9XFQ4 A0A034VF73 A0A182R0W5 B4NNM5 A0A182X6G6 A0A182I3R9 A0A182VMA7 A0A1I8NYK8 V4B577 A0A1W4WT24 A0A2T7PEC0 T1PIC5 B4ME48 A0A182P9W2
Pubmed
19121390
26354079
22118469
24845553
20566863
12364791
+ More
14747013 17210077 21719571 25244985 20798317 30249741 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21347285 22936249 26109357 26109356 24945155 24330624 24508170 24495485 17550304 28004739 17510324 25830018 25542378 18057021 15632085 25348373 23254933
14747013 17210077 21719571 25244985 20798317 30249741 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21347285 22936249 26109357 26109356 24945155 24330624 24508170 24495485 17550304 28004739 17510324 25830018 25542378 18057021 15632085 25348373 23254933
EMBL
BABH01014484
DQ311308
ABD36252.1
HM037181
ADM07446.1
NWSH01002019
+ More
PCG69402.1 PCG69404.1 PCG69403.1 KQ459606 KPI91776.1 KQ461103 KPJ09421.1 AGBW02013516 OWR43311.1 GBYB01013219 JAG82986.1 GBYB01013220 JAG82987.1 KK852458 KDR23583.1 DS235053 EEB11071.1 AAAB01008807 EAA04140.2 GL888087 EGI67740.1 KZ288266 PBC30227.1 KQ435803 KOX73109.1 NEVH01016946 PNF25008.1 GL437918 EFN69859.1 PNF25009.1 KQ414736 KOC62137.1 GL449978 EFN81648.1 GECZ01014061 GECZ01002327 JAS55708.1 JAS67442.1 DS232215 EDS37743.1 GFDL01011869 JAV23176.1 GECZ01006936 GECZ01005220 JAS62833.1 JAS64549.1 GFDL01011765 JAV23280.1 QOIP01000007 RLU21054.1 GECU01020457 JAS87249.1 GFDL01011870 JAV23175.1 GGFJ01008944 MBW58085.1 GECU01037020 JAS70686.1 GEBQ01018747 JAT21230.1 GAMD01002792 JAA98798.1 KQ981285 KYN43183.1 PNF25010.1 CM000362 EDX06312.1 CH480816 EDW47069.1 GGFJ01009314 MBW58455.1 GEBQ01003701 JAT36276.1 AE013599 AAG22295.3 KQ979695 KYN19711.1 ADTU01017084 BT001448 AAN71203.1 CM002911 KMY92476.1 BT083449 AAM68816.2 ACR10181.1 CH954177 KQS70798.1 AAF58970.1 AHN56035.1 AHN56036.1 AB035646 BAA88238.1 GAPW01004125 JAC09473.1 KMY92477.1 KMY92475.1 GALA01001495 JAA93357.1 GEDC01031015 JAS06283.1 KK107152 EZA57077.1 KQ982603 KYQ53959.1 GAMC01004686 JAC01870.1 CM000157 KRJ98467.1 KQS70799.1 GEZM01072774 JAV65402.1 KMY92474.1 AY069328 AAL39473.1 AAM68817.1 AAM68818.1 EDV59401.2 BT003196 AAO24951.1 GANO01002771 JAB57100.1 EDW89473.2 CH477410 EJY57660.1 EAT41483.1 EAT41485.1 KQ434948 KZC12419.1 GEDC01026980 JAS10318.1 CH933808 KRG03987.1 GBXI01015873 GBXI01012123 GBXI01009175 GBXI01007891 GBXI01000710 JAC98418.1 JAD02169.1 JAD05117.1 JAD06401.1 JAD13582.1 KQ978068 KYM97587.1 OUUW01000001 SPP75610.1 GAMC01004685 JAC01871.1 KP216765 AJC97122.1 CH902619 EDV36346.2 KPU76161.1 KPU76162.1 KPU76163.1 CM000071 ENO01823.1 KRG03988.1 GAKP01018503 GAKP01018502 GAKP01018501 JAC40450.1 AXCN02000591 CH964282 EDW85964.2 APCN01000218 KB200149 ESP02671.1 PZQS01000004 PVD31765.1 KA647885 AFP62514.1 CH940662 EDW58813.2
PCG69402.1 PCG69404.1 PCG69403.1 KQ459606 KPI91776.1 KQ461103 KPJ09421.1 AGBW02013516 OWR43311.1 GBYB01013219 JAG82986.1 GBYB01013220 JAG82987.1 KK852458 KDR23583.1 DS235053 EEB11071.1 AAAB01008807 EAA04140.2 GL888087 EGI67740.1 KZ288266 PBC30227.1 KQ435803 KOX73109.1 NEVH01016946 PNF25008.1 GL437918 EFN69859.1 PNF25009.1 KQ414736 KOC62137.1 GL449978 EFN81648.1 GECZ01014061 GECZ01002327 JAS55708.1 JAS67442.1 DS232215 EDS37743.1 GFDL01011869 JAV23176.1 GECZ01006936 GECZ01005220 JAS62833.1 JAS64549.1 GFDL01011765 JAV23280.1 QOIP01000007 RLU21054.1 GECU01020457 JAS87249.1 GFDL01011870 JAV23175.1 GGFJ01008944 MBW58085.1 GECU01037020 JAS70686.1 GEBQ01018747 JAT21230.1 GAMD01002792 JAA98798.1 KQ981285 KYN43183.1 PNF25010.1 CM000362 EDX06312.1 CH480816 EDW47069.1 GGFJ01009314 MBW58455.1 GEBQ01003701 JAT36276.1 AE013599 AAG22295.3 KQ979695 KYN19711.1 ADTU01017084 BT001448 AAN71203.1 CM002911 KMY92476.1 BT083449 AAM68816.2 ACR10181.1 CH954177 KQS70798.1 AAF58970.1 AHN56035.1 AHN56036.1 AB035646 BAA88238.1 GAPW01004125 JAC09473.1 KMY92477.1 KMY92475.1 GALA01001495 JAA93357.1 GEDC01031015 JAS06283.1 KK107152 EZA57077.1 KQ982603 KYQ53959.1 GAMC01004686 JAC01870.1 CM000157 KRJ98467.1 KQS70799.1 GEZM01072774 JAV65402.1 KMY92474.1 AY069328 AAL39473.1 AAM68817.1 AAM68818.1 EDV59401.2 BT003196 AAO24951.1 GANO01002771 JAB57100.1 EDW89473.2 CH477410 EJY57660.1 EAT41483.1 EAT41485.1 KQ434948 KZC12419.1 GEDC01026980 JAS10318.1 CH933808 KRG03987.1 GBXI01015873 GBXI01012123 GBXI01009175 GBXI01007891 GBXI01000710 JAC98418.1 JAD02169.1 JAD05117.1 JAD06401.1 JAD13582.1 KQ978068 KYM97587.1 OUUW01000001 SPP75610.1 GAMC01004685 JAC01871.1 KP216765 AJC97122.1 CH902619 EDV36346.2 KPU76161.1 KPU76162.1 KPU76163.1 CM000071 ENO01823.1 KRG03988.1 GAKP01018503 GAKP01018502 GAKP01018501 JAC40450.1 AXCN02000591 CH964282 EDW85964.2 APCN01000218 KB200149 ESP02671.1 PZQS01000004 PVD31765.1 KA647885 AFP62514.1 CH940662 EDW58813.2
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000005203
+ More
UP000027135 UP000009046 UP000075902 UP000007062 UP000075901 UP000007755 UP000242457 UP000053105 UP000076408 UP000235965 UP000000311 UP000075881 UP000053825 UP000008237 UP000002320 UP000279307 UP000078541 UP000000304 UP000001292 UP000000803 UP000078492 UP000005205 UP000008711 UP000053097 UP000075809 UP000002282 UP000008820 UP000076502 UP000009192 UP000078542 UP000192221 UP000268350 UP000192223 UP000007801 UP000001819 UP000075886 UP000007798 UP000076407 UP000075840 UP000075903 UP000095300 UP000030746 UP000245119 UP000008792 UP000075885
UP000027135 UP000009046 UP000075902 UP000007062 UP000075901 UP000007755 UP000242457 UP000053105 UP000076408 UP000235965 UP000000311 UP000075881 UP000053825 UP000008237 UP000002320 UP000279307 UP000078541 UP000000304 UP000001292 UP000000803 UP000078492 UP000005205 UP000008711 UP000053097 UP000075809 UP000002282 UP000008820 UP000076502 UP000009192 UP000078542 UP000192221 UP000268350 UP000192223 UP000007801 UP000001819 UP000075886 UP000007798 UP000076407 UP000075840 UP000075903 UP000095300 UP000030746 UP000245119 UP000008792 UP000075885
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9IZM6
Q2F5W5
F2VY73
A0A2A4JDA7
A0A2A4JCX7
A0A2A4JDB5
+ More
A0A194PEJ3 A0A194R0Q2 A0A212EPC9 A0A088AJT1 A0A0C9R9Y5 A0A0C9RW95 A0A067RKP3 E0VCG5 A0A182U6P0 Q7QJT8 A0A182S9B0 F4WDG0 A0A2A3EER7 A0A0M9A049 A0A182XZG9 A0A2J7Q8U2 E2A9Q7 A0A182JYQ1 A0A2J7Q8U7 A0A0L7QTZ9 E2BRQ4 A0A1B6FZT9 B0WZR8 A0A1Q3F6J3 A0A1B6GKK6 A0A1Q3F6U4 A0A3L8DKT4 A0A1B6IK06 A0A1Q3F6I0 A0A2M4BYC1 A0A1B6H7K9 A0A1B6LC41 T1E7S6 A0A195FRW0 A0A2J7Q8V1 B4QGT7 B4HSP2 A0A2M4BZF2 A0A1B6MK13 A1Z7S2 A0A151J7K8 A0A158NIR4 Q8IH37 A0A0J9R8X9 A1Z7S1 A0A0Q5WKM2 A1Z7S3 A0A0B4LEY0 Q7KY07 A0A023EK20 A0A0J9R8T1 A0A0J9TX72 T1E268 A0A1B6BYD4 A0A026WLZ2 A0A151X0T5 W8CBL4 A0A0R1DTY3 A0A0Q5WNC0 A0A1Y1KVJ8 A0A0J9R9W6 Q8T0G8 B3N7J6 Q86PD0 U5ERJ9 B4P3Z0 J9HYU4 Q174H5 A0A154PKM5 A0A1B6CA05 A0A0Q9XJX9 A0A0A1XRI0 A0A195C9U3 A0A1W4UY09 A0A3B0JJQ9 W8BKL3 A0A0B4UBW9 A0A1W4X2P4 B3MD31 N6W6G8 A0A0Q9XFQ4 A0A034VF73 A0A182R0W5 B4NNM5 A0A182X6G6 A0A182I3R9 A0A182VMA7 A0A1I8NYK8 V4B577 A0A1W4WT24 A0A2T7PEC0 T1PIC5 B4ME48 A0A182P9W2
A0A194PEJ3 A0A194R0Q2 A0A212EPC9 A0A088AJT1 A0A0C9R9Y5 A0A0C9RW95 A0A067RKP3 E0VCG5 A0A182U6P0 Q7QJT8 A0A182S9B0 F4WDG0 A0A2A3EER7 A0A0M9A049 A0A182XZG9 A0A2J7Q8U2 E2A9Q7 A0A182JYQ1 A0A2J7Q8U7 A0A0L7QTZ9 E2BRQ4 A0A1B6FZT9 B0WZR8 A0A1Q3F6J3 A0A1B6GKK6 A0A1Q3F6U4 A0A3L8DKT4 A0A1B6IK06 A0A1Q3F6I0 A0A2M4BYC1 A0A1B6H7K9 A0A1B6LC41 T1E7S6 A0A195FRW0 A0A2J7Q8V1 B4QGT7 B4HSP2 A0A2M4BZF2 A0A1B6MK13 A1Z7S2 A0A151J7K8 A0A158NIR4 Q8IH37 A0A0J9R8X9 A1Z7S1 A0A0Q5WKM2 A1Z7S3 A0A0B4LEY0 Q7KY07 A0A023EK20 A0A0J9R8T1 A0A0J9TX72 T1E268 A0A1B6BYD4 A0A026WLZ2 A0A151X0T5 W8CBL4 A0A0R1DTY3 A0A0Q5WNC0 A0A1Y1KVJ8 A0A0J9R9W6 Q8T0G8 B3N7J6 Q86PD0 U5ERJ9 B4P3Z0 J9HYU4 Q174H5 A0A154PKM5 A0A1B6CA05 A0A0Q9XJX9 A0A0A1XRI0 A0A195C9U3 A0A1W4UY09 A0A3B0JJQ9 W8BKL3 A0A0B4UBW9 A0A1W4X2P4 B3MD31 N6W6G8 A0A0Q9XFQ4 A0A034VF73 A0A182R0W5 B4NNM5 A0A182X6G6 A0A182I3R9 A0A182VMA7 A0A1I8NYK8 V4B577 A0A1W4WT24 A0A2T7PEC0 T1PIC5 B4ME48 A0A182P9W2
PDB
4CZ2
E-value=1.45218e-82,
Score=777
Ontologies
GO
GO:0003924
GO:0016192
GO:0005802
GO:0031982
GO:0005525
GO:0032482
GO:0005739
GO:0042470
GO:0006886
GO:0016021
GO:0006727
GO:0005776
GO:0010506
GO:0048072
GO:0010883
GO:0005764
GO:0008057
GO:0055037
GO:0006856
GO:0045202
GO:1901214
GO:0007264
GO:0016020
GO:0003995
GO:0003997
GO:0005777
GO:0006631
GO:0006635
GO:0006281
GO:0048384
GO:0005794
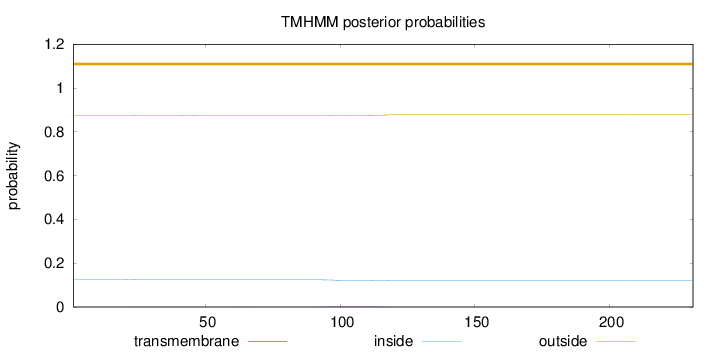
Topology
Length:
231
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10526
Exp number, first 60 AAs:
0.01167
Total prob of N-in:
0.12617
outside
1 - 231
Population Genetic Test Statistics
Pi
210.258562
Theta
171.521093
Tajima's D
0.747681
CLR
0.022104
CSRT
0.587020648967552
Interpretation
Uncertain
