Gene
KWMTBOMO02494
Pre Gene Modal
BGIBMGA002712
Annotation
protein_white-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.317
Sequence
CDS
ATGGAGTTGAATAAGGTCCTCTTAGACGAATTCGAAAATAAAGACGCAATTATTGTTAGAAATCTGAAGGTTTGGACGCCAGAAGAGAAGTCAATATGGAGAAAAGTTGCTAAACCAAAAACTGTGATAATCGATAATGTTTCAGCGTGTATAAGGGAAGGGGAATTTGCAGCAATCATCGGGCCGAGTGGAGCAGGAAAAACAACATTTCTAGTGTCGTTAGCCGGGAAATGCACATTACCATTTGAAGGCACCGTTACGATCAACGGTAGAAATGTCAGGGATCTGACGGGTGCCGAGATCGTGCCCCAGTTCGACGTGTTCACGGACAGCTTGACTGTTATGGAGCAACTTGTTTTTATGACAGAAATGAAACTAGGTAACAGTACGAAGCAACCGAACAAATCAATTCTGAATACAGTCATCGAAGAATTCAAACTAAGCGCACACGTTGAAACGCGCATAGGATCATTATCTGGCGGAGAAAGGAGGTTGTTGTCTTTAGCCACTTCGTTCTTATCGAATCCACAAATATTGATATGCGACGAACCCACTACCGGCTTGGATAGTTATAACGCATCACAAGTAATTGGCATATTAAAGAAATTGTCAGCGTCCGGTAAGATTGTCATATGTTCTGTTCATCAACCTTCGTGTGATATCTTCAAAGAGTTTAACTCTATTCTTTTGATGGCTGAGGGAAATCTACTTTTCCATGGAACTCAGGATGCGTGCAAATCGTTCTTTGAAAGCATTGACCTGCATTGCCCGTTGAACTATAATCCAGCAGAATTTTACATACGAGCTGTATCCAATCACAATGGAGTATGTATCAAAAAGATGCTGGAAAATTACCAGGACCAGTCTTTGGACTGTGACGAAACCGGTTTGGAAGTTAACCTAATCTATAAAAAGCCACAAAGAAATTGGCTGAAGCAAGTGCAACTTCTACTGTGGAGGTCTTCTTTGAGCCTGAAGAGCGATTTAAAAAGTTACGTGTTTCAACTGCTCTTGTCTGTGGTAGTAACAGCATCAGTTCTTGGCACTGTCTACAGCGGAGTCACAGGTACCACACAACGTGGAATTCAGGACGTCAGAGGCTTCCTTTGGTTGGTCACGAGCGAAGTTTGTTACGGGCTTGCCTATAGCACCCTCTATGTGTTCAAATATGAAGTCACTCTGTTCAGGAGAGAAGTCGGCATGTACAAATGCTCAGCATATTTCGTTAGCAAATTTTTGAGTTTTATACCAAGGTGCGTTATCTGGCCCATCGCACTGGTGTGTACTACAACCGTTGCGATCGAGTTACCAAACCACGTAATCACCACGATGGAATTTATTGTCGTGCTGATATTTGCTGCGATTGCTTCCATGGCTTATGGTCTCGGTATGTCTGCATTGTTTACATCGACAGGCAACATGGGCGACGTGATGCCTTGCTTCGATCTTCCATTGTTTCTAATGTCAGGAGCATTCCTAACGATCTCAACGTTACCAATATGGTTATATCCCGTGAAATTCATCTCGCACTTCTACTACGCCATGGATACTATCAGCAATCTTTACTGGAGACAGATTCTTTATATTGATTGTCCAGTGAACACAACTACGACGTGCACGAGTAGCGGAGAAGCAGTATTGTACGAAATTGGATATTCTAATAATTTTGTACTACAAAACAGTTTGGGCTTATTACTGGTTACTACTATGTGGGGCTTACTGGGTTATTACGGGATGAAACGAGAAGAGAAAAAGGGTTACGCTTACTGA
Protein
MELNKVLLDEFENKDAIIVRNLKVWTPEEKSIWRKVAKPKTVIIDNVSACIREGEFAAIIGPSGAGKTTFLVSLAGKCTLPFEGTVTINGRNVRDLTGAEIVPQFDVFTDSLTVMEQLVFMTEMKLGNSTKQPNKSILNTVIEEFKLSAHVETRIGSLSGGERRLLSLATSFLSNPQILICDEPTTGLDSYNASQVIGILKKLSASGKIVICSVHQPSCDIFKEFNSILLMAEGNLLFHGTQDACKSFFESIDLHCPLNYNPAEFYIRAVSNHNGVCIKKMLENYQDQSLDCDETGLEVNLIYKKPQRNWLKQVQLLLWRSSLSLKSDLKSYVFQLLLSVVVTASVLGTVYSGVTGTTQRGIQDVRGFLWLVTSEVCYGLAYSTLYVFKYEVTLFRREVGMYKCSAYFVSKFLSFIPRCVIWPIALVCTTTVAIELPNHVITTMEFIVVLIFAAIASMAYGLGMSALFTSTGNMGDVMPCFDLPLFLMSGAFLTISTLPIWLYPVKFISHFYYAMDTISNLYWRQILYIDCPVNTTTTCTSSGEAVLYEIGYSNNFVLQNSLGLLLVTTMWGLLGYYGMKREEKKGYAY
Summary
Uniprot
S6BR83
S6BBX8
S6B597
S6BKX7
A0A1C8V605
A0A194PKZ3
+ More
A0A0N9DUU6 A0A2H1VR06 A0A212F844 E2A9Q8 A0A0K0LT29 A0A1B6FJA2 A0A154PMA4 A0A288W7L2 A0A2P2I4T6 A0A1B6DHK4 A0A212F822 A0A336KXP5 D6X2N7 A0A146KYL8 E0VCR4 A0A2S2NJ69 N6USD5 A0A1J1J8K5 A0A3Q0IQI1 J9K9D4 T1HEX6 U4UAU9 A0A132AHL9 A0A0P4ZJR1 A0A0P4ZAD0 A0A0P5VFK7 A0A0P5U388 A0A0N8AYK3 A0A0P4YQX3 A0A0N7ZSY2 E9GAJ6 A0A0P5VA46 A0A0P5V3Q8 A0A0P6ECT1 A0A0P6HM62 A0A0P5NWW5 A0A0N8CM25 A0A0P6CJI3 A0A0P6FXN1 A0A0P5U3A1 A0A0N8C292 A0A0P4YYV1 A0A0P5IHJ5 A0A0P5A6M0 A0A0P5KG52 A0A0P5NWT5 A0A0P5JC00 A0A0P6EXU0 A0A0P5VBE7 A0A0P5Q224 A0A0P5QGG4 A0A0P5DD44 A0A0P6D5W5 A0A0P5R7U2 A0A0P5X0P4 A0A0P6CJ67 A0A0P5SEL1 A0A0P5L8X2 A0A0P5X0A4 A0A0P5RR57 A0A0P5QEE3 A0A0P6GL71 A0A0N8CQV2 A0A0P5XPA5 A0A0P5UPU4 A0A0P6GVL9 A0A0P6CMG5 A0A0P5MIA2 A0A0P5ILM3 A0A162EDY8 A0A0N8D089 A0A0N7ZS33 A0A0P5JEF2 A0A0P5JZ26 A0A0N8CKN8 A0A0N8EAE4 A0A088AJS3 A0A0P5R8T7 A0A0P5PXA3 A0A0P6FGP4 A0A0P5XVW2 A0A0P5HK00 A0A0P6HCD3 A0A0P4XUU7 A0A0P5WA77 A0A0P6DLW9 A0A0P5WUM5 A0A0P5PKJ3 A0A0P4Y0Q5
A0A0N9DUU6 A0A2H1VR06 A0A212F844 E2A9Q8 A0A0K0LT29 A0A1B6FJA2 A0A154PMA4 A0A288W7L2 A0A2P2I4T6 A0A1B6DHK4 A0A212F822 A0A336KXP5 D6X2N7 A0A146KYL8 E0VCR4 A0A2S2NJ69 N6USD5 A0A1J1J8K5 A0A3Q0IQI1 J9K9D4 T1HEX6 U4UAU9 A0A132AHL9 A0A0P4ZJR1 A0A0P4ZAD0 A0A0P5VFK7 A0A0P5U388 A0A0N8AYK3 A0A0P4YQX3 A0A0N7ZSY2 E9GAJ6 A0A0P5VA46 A0A0P5V3Q8 A0A0P6ECT1 A0A0P6HM62 A0A0P5NWW5 A0A0N8CM25 A0A0P6CJI3 A0A0P6FXN1 A0A0P5U3A1 A0A0N8C292 A0A0P4YYV1 A0A0P5IHJ5 A0A0P5A6M0 A0A0P5KG52 A0A0P5NWT5 A0A0P5JC00 A0A0P6EXU0 A0A0P5VBE7 A0A0P5Q224 A0A0P5QGG4 A0A0P5DD44 A0A0P6D5W5 A0A0P5R7U2 A0A0P5X0P4 A0A0P6CJ67 A0A0P5SEL1 A0A0P5L8X2 A0A0P5X0A4 A0A0P5RR57 A0A0P5QEE3 A0A0P6GL71 A0A0N8CQV2 A0A0P5XPA5 A0A0P5UPU4 A0A0P6GVL9 A0A0P6CMG5 A0A0P5MIA2 A0A0P5ILM3 A0A162EDY8 A0A0N8D089 A0A0N7ZS33 A0A0P5JEF2 A0A0P5JZ26 A0A0N8CKN8 A0A0N8EAE4 A0A088AJS3 A0A0P5R8T7 A0A0P5PXA3 A0A0P6FGP4 A0A0P5XVW2 A0A0P5HK00 A0A0P6HCD3 A0A0P4XUU7 A0A0P5WA77 A0A0P6DLW9 A0A0P5WUM5 A0A0P5PKJ3 A0A0P4Y0Q5
Pubmed
EMBL
AB780447
BAN66702.1
AB780446
BAN66701.1
AB780448
BAN66703.1
+ More
AB780449 BAN66704.1 KU754480 ANW09742.1 KQ459606 KPI91775.1 KT781081 ALF12136.1 ODYU01003887 SOQ43206.1 AGBW02009802 OWR49891.1 GL437918 EFN69860.1 KP120764 KQ971307 AJD07061.1 KYB29666.1 GECZ01019500 JAS50269.1 KQ434948 KZC12418.1 KX880378 ARS88259.1 IACF01003431 LAB69047.1 GEDC01027124 GEDC01012117 JAS10174.1 JAS25181.1 OWR49892.1 UFQS01001196 UFQT01001196 SSX09640.1 SSX29436.1 KQ971372 EFA09850.1 GDHC01017351 GDHC01005996 JAQ01278.1 JAQ12633.1 DS235063 EEB11170.1 GGMR01004598 MBY17217.1 APGK01018646 APGK01018647 KB740081 ENN81632.1 CVRI01000075 CRL08298.1 ABLF02032635 ABLF02032639 ABLF02032642 ACPB03020694 KB632220 ERL90182.1 JXLN01015229 KPM10492.1 GDIP01224974 JAI98427.1 GDIP01215678 JAJ07724.1 GDIP01102392 GDIP01102391 JAM01323.1 GDIP01118451 JAL85263.1 GDIQ01230235 JAK21490.1 GDIP01224975 JAI98426.1 GDIP01215681 JAJ07721.1 GL732537 EFX83518.1 GDIP01102395 JAM01320.1 GDIP01105233 JAL98481.1 GDIQ01233739 GDIQ01141133 GDIQ01064563 GDIQ01043855 GDIQ01041416 GDIQ01038424 JAN30174.1 GDIQ01017488 JAN77249.1 GDIQ01136623 JAL15103.1 GDIP01118447 JAL85267.1 GDIQ01090395 JAN04342.1 GDIQ01059166 JAN35571.1 GDIP01118448 JAL85266.1 GDIQ01126037 GDIQ01121923 JAL29803.1 GDIP01224973 GDIP01215677 JAI98428.1 GDIQ01212389 JAK39336.1 GDIP01215676 JAJ07726.1 GDIQ01184668 JAK67057.1 GDIQ01136625 GDIP01120540 GDIP01107783 GDIQ01090394 JAL15101.1 JAL83174.1 GDIQ01202296 GDIQ01202295 GDIQ01202294 JAK49429.1 GDIP01120541 GDIQ01056509 JAL83173.1 JAN38228.1 GDIP01188573 GDIP01102432 GDIP01102393 JAM01322.1 GDIQ01141118 JAL10608.1 GDIQ01124269 JAL27457.1 GDIP01159290 JAJ64112.1 GDIQ01090389 JAN04348.1 GDIQ01124268 JAL27458.1 GDIP01079092 JAM24623.1 GDIQ01090392 JAN04345.1 GDIQ01092839 JAL58887.1 GDIQ01173105 GDIQ01173104 JAK78620.1 GDIP01079093 JAM24622.1 GDIQ01097183 JAL54543.1 GDIQ01136620 GDIQ01097182 GDIQ01097181 GDIQ01046115 JAL15106.1 GDIQ01033007 JAN61730.1 GDIQ01197003 GDIP01120544 GDIP01107831 JAK54722.1 JAL95883.1 GDIP01081536 JAM22179.1 GDIP01109990 JAL93724.1 GDIQ01028874 JAN65863.1 GDIQ01090391 JAN04346.1 GDIQ01157720 JAK94005.1 GDIQ01212391 GDIQ01212390 JAK39335.1 LRGB01002019 KZS09610.1 GDIP01081535 JAM22180.1 GDIP01229483 GDIP01218073 GDIP01083452 JAJ05329.1 GDIQ01200762 JAK50963.1 GDIQ01197004 JAK54721.1 GDIP01122343 JAL81371.1 GDIQ01160158 GDIQ01157719 GDIQ01046116 JAN48621.1 GDIQ01217290 GDIQ01110981 JAL40745.1 GDIQ01142919 GDIP01069799 JAL08807.1 JAM33916.1 GDIQ01251389 GDIQ01249886 GDIQ01237247 GDIQ01196697 GDIQ01187908 GDIQ01171675 GDIQ01143126 GDIQ01047614 JAN47123.1 KZS09608.1 GDIP01079094 JAM24621.1 GDIQ01232277 GDIQ01142918 JAK19448.1 GDIQ01061949 GDIQ01032167 GDIQ01030806 JAN63931.1 GDIP01236315 JAI87086.1 GDIP01090186 JAM13529.1 GDIQ01090390 JAN04347.1 GDIP01081537 JAM22178.1 GDIQ01127629 JAL24097.1 GDIP01236316 JAI87085.1
AB780449 BAN66704.1 KU754480 ANW09742.1 KQ459606 KPI91775.1 KT781081 ALF12136.1 ODYU01003887 SOQ43206.1 AGBW02009802 OWR49891.1 GL437918 EFN69860.1 KP120764 KQ971307 AJD07061.1 KYB29666.1 GECZ01019500 JAS50269.1 KQ434948 KZC12418.1 KX880378 ARS88259.1 IACF01003431 LAB69047.1 GEDC01027124 GEDC01012117 JAS10174.1 JAS25181.1 OWR49892.1 UFQS01001196 UFQT01001196 SSX09640.1 SSX29436.1 KQ971372 EFA09850.1 GDHC01017351 GDHC01005996 JAQ01278.1 JAQ12633.1 DS235063 EEB11170.1 GGMR01004598 MBY17217.1 APGK01018646 APGK01018647 KB740081 ENN81632.1 CVRI01000075 CRL08298.1 ABLF02032635 ABLF02032639 ABLF02032642 ACPB03020694 KB632220 ERL90182.1 JXLN01015229 KPM10492.1 GDIP01224974 JAI98427.1 GDIP01215678 JAJ07724.1 GDIP01102392 GDIP01102391 JAM01323.1 GDIP01118451 JAL85263.1 GDIQ01230235 JAK21490.1 GDIP01224975 JAI98426.1 GDIP01215681 JAJ07721.1 GL732537 EFX83518.1 GDIP01102395 JAM01320.1 GDIP01105233 JAL98481.1 GDIQ01233739 GDIQ01141133 GDIQ01064563 GDIQ01043855 GDIQ01041416 GDIQ01038424 JAN30174.1 GDIQ01017488 JAN77249.1 GDIQ01136623 JAL15103.1 GDIP01118447 JAL85267.1 GDIQ01090395 JAN04342.1 GDIQ01059166 JAN35571.1 GDIP01118448 JAL85266.1 GDIQ01126037 GDIQ01121923 JAL29803.1 GDIP01224973 GDIP01215677 JAI98428.1 GDIQ01212389 JAK39336.1 GDIP01215676 JAJ07726.1 GDIQ01184668 JAK67057.1 GDIQ01136625 GDIP01120540 GDIP01107783 GDIQ01090394 JAL15101.1 JAL83174.1 GDIQ01202296 GDIQ01202295 GDIQ01202294 JAK49429.1 GDIP01120541 GDIQ01056509 JAL83173.1 JAN38228.1 GDIP01188573 GDIP01102432 GDIP01102393 JAM01322.1 GDIQ01141118 JAL10608.1 GDIQ01124269 JAL27457.1 GDIP01159290 JAJ64112.1 GDIQ01090389 JAN04348.1 GDIQ01124268 JAL27458.1 GDIP01079092 JAM24623.1 GDIQ01090392 JAN04345.1 GDIQ01092839 JAL58887.1 GDIQ01173105 GDIQ01173104 JAK78620.1 GDIP01079093 JAM24622.1 GDIQ01097183 JAL54543.1 GDIQ01136620 GDIQ01097182 GDIQ01097181 GDIQ01046115 JAL15106.1 GDIQ01033007 JAN61730.1 GDIQ01197003 GDIP01120544 GDIP01107831 JAK54722.1 JAL95883.1 GDIP01081536 JAM22179.1 GDIP01109990 JAL93724.1 GDIQ01028874 JAN65863.1 GDIQ01090391 JAN04346.1 GDIQ01157720 JAK94005.1 GDIQ01212391 GDIQ01212390 JAK39335.1 LRGB01002019 KZS09610.1 GDIP01081535 JAM22180.1 GDIP01229483 GDIP01218073 GDIP01083452 JAJ05329.1 GDIQ01200762 JAK50963.1 GDIQ01197004 JAK54721.1 GDIP01122343 JAL81371.1 GDIQ01160158 GDIQ01157719 GDIQ01046116 JAN48621.1 GDIQ01217290 GDIQ01110981 JAL40745.1 GDIQ01142919 GDIP01069799 JAL08807.1 JAM33916.1 GDIQ01251389 GDIQ01249886 GDIQ01237247 GDIQ01196697 GDIQ01187908 GDIQ01171675 GDIQ01143126 GDIQ01047614 JAN47123.1 KZS09608.1 GDIP01079094 JAM24621.1 GDIQ01232277 GDIQ01142918 JAK19448.1 GDIQ01061949 GDIQ01032167 GDIQ01030806 JAN63931.1 GDIP01236315 JAI87086.1 GDIP01090186 JAM13529.1 GDIQ01090390 JAN04347.1 GDIP01081537 JAM22178.1 GDIQ01127629 JAL24097.1 GDIP01236316 JAI87085.1
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
S6BR83
S6BBX8
S6B597
S6BKX7
A0A1C8V605
A0A194PKZ3
+ More
A0A0N9DUU6 A0A2H1VR06 A0A212F844 E2A9Q8 A0A0K0LT29 A0A1B6FJA2 A0A154PMA4 A0A288W7L2 A0A2P2I4T6 A0A1B6DHK4 A0A212F822 A0A336KXP5 D6X2N7 A0A146KYL8 E0VCR4 A0A2S2NJ69 N6USD5 A0A1J1J8K5 A0A3Q0IQI1 J9K9D4 T1HEX6 U4UAU9 A0A132AHL9 A0A0P4ZJR1 A0A0P4ZAD0 A0A0P5VFK7 A0A0P5U388 A0A0N8AYK3 A0A0P4YQX3 A0A0N7ZSY2 E9GAJ6 A0A0P5VA46 A0A0P5V3Q8 A0A0P6ECT1 A0A0P6HM62 A0A0P5NWW5 A0A0N8CM25 A0A0P6CJI3 A0A0P6FXN1 A0A0P5U3A1 A0A0N8C292 A0A0P4YYV1 A0A0P5IHJ5 A0A0P5A6M0 A0A0P5KG52 A0A0P5NWT5 A0A0P5JC00 A0A0P6EXU0 A0A0P5VBE7 A0A0P5Q224 A0A0P5QGG4 A0A0P5DD44 A0A0P6D5W5 A0A0P5R7U2 A0A0P5X0P4 A0A0P6CJ67 A0A0P5SEL1 A0A0P5L8X2 A0A0P5X0A4 A0A0P5RR57 A0A0P5QEE3 A0A0P6GL71 A0A0N8CQV2 A0A0P5XPA5 A0A0P5UPU4 A0A0P6GVL9 A0A0P6CMG5 A0A0P5MIA2 A0A0P5ILM3 A0A162EDY8 A0A0N8D089 A0A0N7ZS33 A0A0P5JEF2 A0A0P5JZ26 A0A0N8CKN8 A0A0N8EAE4 A0A088AJS3 A0A0P5R8T7 A0A0P5PXA3 A0A0P6FGP4 A0A0P5XVW2 A0A0P5HK00 A0A0P6HCD3 A0A0P4XUU7 A0A0P5WA77 A0A0P6DLW9 A0A0P5WUM5 A0A0P5PKJ3 A0A0P4Y0Q5
A0A0N9DUU6 A0A2H1VR06 A0A212F844 E2A9Q8 A0A0K0LT29 A0A1B6FJA2 A0A154PMA4 A0A288W7L2 A0A2P2I4T6 A0A1B6DHK4 A0A212F822 A0A336KXP5 D6X2N7 A0A146KYL8 E0VCR4 A0A2S2NJ69 N6USD5 A0A1J1J8K5 A0A3Q0IQI1 J9K9D4 T1HEX6 U4UAU9 A0A132AHL9 A0A0P4ZJR1 A0A0P4ZAD0 A0A0P5VFK7 A0A0P5U388 A0A0N8AYK3 A0A0P4YQX3 A0A0N7ZSY2 E9GAJ6 A0A0P5VA46 A0A0P5V3Q8 A0A0P6ECT1 A0A0P6HM62 A0A0P5NWW5 A0A0N8CM25 A0A0P6CJI3 A0A0P6FXN1 A0A0P5U3A1 A0A0N8C292 A0A0P4YYV1 A0A0P5IHJ5 A0A0P5A6M0 A0A0P5KG52 A0A0P5NWT5 A0A0P5JC00 A0A0P6EXU0 A0A0P5VBE7 A0A0P5Q224 A0A0P5QGG4 A0A0P5DD44 A0A0P6D5W5 A0A0P5R7U2 A0A0P5X0P4 A0A0P6CJ67 A0A0P5SEL1 A0A0P5L8X2 A0A0P5X0A4 A0A0P5RR57 A0A0P5QEE3 A0A0P6GL71 A0A0N8CQV2 A0A0P5XPA5 A0A0P5UPU4 A0A0P6GVL9 A0A0P6CMG5 A0A0P5MIA2 A0A0P5ILM3 A0A162EDY8 A0A0N8D089 A0A0N7ZS33 A0A0P5JEF2 A0A0P5JZ26 A0A0N8CKN8 A0A0N8EAE4 A0A088AJS3 A0A0P5R8T7 A0A0P5PXA3 A0A0P6FGP4 A0A0P5XVW2 A0A0P5HK00 A0A0P6HCD3 A0A0P4XUU7 A0A0P5WA77 A0A0P6DLW9 A0A0P5WUM5 A0A0P5PKJ3 A0A0P4Y0Q5
PDB
6FFC
E-value=1.31719e-28,
Score=316
Ontologies
GO
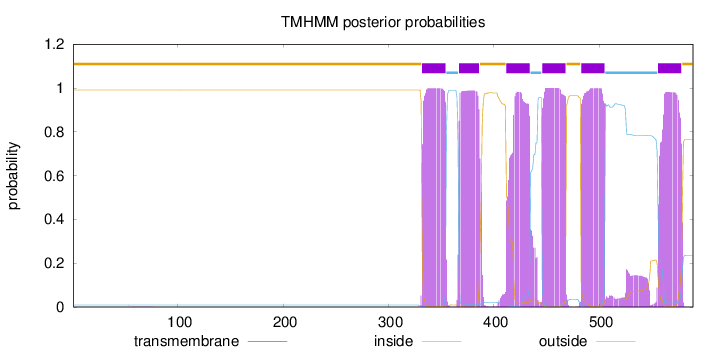
Topology
Length:
589
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
136.56464
Exp number, first 60 AAs:
0.00165
Total prob of N-in:
0.00884
outside
1 - 331
TMhelix
332 - 354
inside
355 - 366
TMhelix
367 - 386
outside
387 - 411
TMhelix
412 - 434
inside
435 - 445
TMhelix
446 - 468
outside
469 - 482
TMhelix
483 - 505
inside
506 - 555
TMhelix
556 - 578
outside
579 - 589
Population Genetic Test Statistics
Pi
238.182424
Theta
174.712807
Tajima's D
1.087458
CLR
0.720363
CSRT
0.68311584420779
Interpretation
Uncertain
