Gene
KWMTBOMO02492
Pre Gene Modal
BGIBMGA002580
Annotation
Bm-ok_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.289 Mitochondrial Reliability : 1.377
Sequence
CDS
ATGAAAGAATACAATCTTAAGCTACCCACGCGCAATGTCCTCACCGGAGCCGTGGTCAACTGGGAGGACGACGAAATGGGAGTGATGGGAGATAGCCCGGAGAATCTGACATTAGCTTGGAAGGACTTATCTGTGTTTAGAAAGAAGAAGATACACACTAGCATGTGGAGATCGGCGGTCTATGAAGAGGTTAAAGTGTTGCATGGAGTAAGCGGCAGTGTCTCGTCCGGAAATCTGGTGGCGTTAATGGGTTCAAGCGGGGCCGGAAAAACCACACTCTTGGCCGCCATCAGCCGTCGTGACAAGAGCGCTCTGACAGGCTACCTCATGCTAAATGGAAGATTAGCCGGCGCTGACCTGATCGCCAGGATCTCAGGCTTCGTGCCTCAGGAGGACTTGGCAATCGAAGATCTCACCGTAGCTGAGCATATGGAGTTCATGGTGAGTCTAATTATGAAGTGA
Protein
MKEYNLKLPTRNVLTGAVVNWEDDEMGVMGDSPENLTLAWKDLSVFRKKKIHTSMWRSAVYEEVKVLHGVSGSVSSGNLVALMGSSGAGKTTLLAAISRRDKSALTGYLMLNGRLAGADLIARISGFVPQEDLAIEDLTVAEHMEFMVSLIMK
Summary
Uniprot
H9IZ95
S6CBY2
S6BKX3
S6BR80
A0A2H1VUG9
A0A2A4IVY0
+ More
A0A1C8V608 A0A1C8V604 A0A212F822 A0A194QV63 A0A194PGE3 A0A1C8V610 A0A1C8V606 A0A0C9RQR3 A0A161MGQ8 A0A1B6HNQ6 A0A0L7KV61 A0A1B6EAM8 A0A2J7RHZ8 A0A2J7RHY9 A0A2J7RHY7 A0A1B6G8L1 A0A088AJS3 A0A2Z5ZAT2 A0A0L7QU96 A0A3Q0J7W2 A0A3Q0J7Y4 A0A1B6M3P2 A0A154PMA4 A0A3Q0IZ92 A0A0A9X9P7 A0A146LTS6 A0A0A9Y4T5 A0A0A9YCE5 A0A0K8SXT9 A0A0N0BFB7 A0A146KYL8 A0A0K8T4R4 A0A0A9Y0I9 A0A2S0X9Z5 A0A0K8T4T1 A0A2H8U0W7 A0A067RUR8 A0A2S2PFL4 A0A0K8V2T9 A0A2S2NKJ3 A0A0K8T554 A0A0K8W3H6 A0A2S2NGC5 J9JZD3 A0A336KXP5 A0A034VX06 J9M761 A0A2H4TET2 A0A1S3DS16 A0A0A1WSU4 B4LMD8 A0A1B6JFE2 A0A1B6K9B6 T1HEX6 A0A2S2RAY9 X1WJG5 A0A2S2R7P5 W8BR39 J9K443 A0A1B6FJA2 A0A1B6L4R2 A0A0M3QV28 A0A1S4G5F6 A0A1B0CN52 A0A0L0C1S9 B3MFG3 A0A2P2I4T6 A0A1I8NA44 A0A2H8TX38 B4KSG8 A0A2P8Z745 B4NMQ3 A0A1I8QE22 A0A1B0ABN1 A0A1B0FJ32 A0A1A9VCA6 A0A2H4TES7 Q867Z8 A0A034W8J1 A0A1I8MRT6 A0A0K8WJK6 E0VCR4 A0A0K8U6H0 B0WZR9 A0A0P5XY92
A0A1C8V608 A0A1C8V604 A0A212F822 A0A194QV63 A0A194PGE3 A0A1C8V610 A0A1C8V606 A0A0C9RQR3 A0A161MGQ8 A0A1B6HNQ6 A0A0L7KV61 A0A1B6EAM8 A0A2J7RHZ8 A0A2J7RHY9 A0A2J7RHY7 A0A1B6G8L1 A0A088AJS3 A0A2Z5ZAT2 A0A0L7QU96 A0A3Q0J7W2 A0A3Q0J7Y4 A0A1B6M3P2 A0A154PMA4 A0A3Q0IZ92 A0A0A9X9P7 A0A146LTS6 A0A0A9Y4T5 A0A0A9YCE5 A0A0K8SXT9 A0A0N0BFB7 A0A146KYL8 A0A0K8T4R4 A0A0A9Y0I9 A0A2S0X9Z5 A0A0K8T4T1 A0A2H8U0W7 A0A067RUR8 A0A2S2PFL4 A0A0K8V2T9 A0A2S2NKJ3 A0A0K8T554 A0A0K8W3H6 A0A2S2NGC5 J9JZD3 A0A336KXP5 A0A034VX06 J9M761 A0A2H4TET2 A0A1S3DS16 A0A0A1WSU4 B4LMD8 A0A1B6JFE2 A0A1B6K9B6 T1HEX6 A0A2S2RAY9 X1WJG5 A0A2S2R7P5 W8BR39 J9K443 A0A1B6FJA2 A0A1B6L4R2 A0A0M3QV28 A0A1S4G5F6 A0A1B0CN52 A0A0L0C1S9 B3MFG3 A0A2P2I4T6 A0A1I8NA44 A0A2H8TX38 B4KSG8 A0A2P8Z745 B4NMQ3 A0A1I8QE22 A0A1B0ABN1 A0A1B0FJ32 A0A1A9VCA6 A0A2H4TES7 Q867Z8 A0A034W8J1 A0A1I8MRT6 A0A0K8WJK6 E0VCR4 A0A0K8U6H0 B0WZR9 A0A0P5XY92
Pubmed
EMBL
BABH01014489
AB780443
BAN66700.1
AB780442
BAN66699.1
AB780440
+ More
BAN66697.1 ODYU01004514 SOQ44475.1 NWSH01006605 PCG63342.1 KU754482 ANW09744.1 KU754481 ANW09743.1 AGBW02009802 OWR49892.1 KQ461103 KPJ09423.1 KQ459606 KPI91774.1 KU754486 ANW09748.1 KU754485 ANW09747.1 GBYB01010845 JAG80612.1 GEMB01002843 JAS00356.1 GECU01031403 GECU01013715 JAS76303.1 JAS93991.1 JTDY01005474 KOB66961.1 GEDC01002338 JAS34960.1 NEVH01003510 PNF40449.1 PNF40448.1 PNF40447.1 GECZ01010984 JAS58785.1 LC368081 BBC77940.1 KQ414736 KOC62134.1 GEBQ01009425 JAT30552.1 KQ434948 KZC12418.1 GBHO01029779 JAG13825.1 GDHC01007506 JAQ11123.1 GBHO01016420 JAG27184.1 GBHO01016419 JAG27185.1 GBRD01008091 GDHC01011007 JAG57730.1 JAQ07622.1 KQ435803 KOX73112.1 GDHC01017351 GDHC01005996 JAQ01278.1 JAQ12633.1 GBRD01005302 JAG60519.1 GBHO01020554 GBRD01005165 JAG23050.1 JAG60656.1 MH105807 AWC08637.1 GBRD01005303 JAG60518.1 GFXV01007876 MBW19681.1 KK852458 KDR23584.1 GGMR01015576 MBY28195.1 GDHF01019143 JAI33171.1 GGMR01004833 MBY17452.1 GBRD01005164 GDHC01016572 JAG60657.1 JAQ02057.1 GDHF01006670 JAI45644.1 GGMR01003601 MBY16220.1 ABLF02038190 UFQS01001196 UFQT01001196 SSX09640.1 SSX29436.1 GAKP01011111 JAC47841.1 ABLF02024643 KY849648 ATY74528.1 GBXI01012586 JAD01706.1 CH940648 EDW62033.1 GECU01009884 JAS97822.1 GEBQ01031928 JAT08049.1 ACPB03020694 GGMS01017627 MBY86830.1 ABLF02024865 GGMS01016781 MBY85984.1 GAMC01010859 JAB95696.1 GECZ01019500 JAS50269.1 GEBQ01021268 JAT18709.1 CP012524 ALC41689.1 AJWK01019808 JRES01000997 KNC26260.1 CH902619 EDV36648.1 IACF01003431 LAB69047.1 GFXV01007001 MBW18806.1 CH933808 EDW09473.1 PYGN01000167 PSN52318.1 CH964282 EDW85642.1 CCAG010001084 KY849652 ATY74530.1 AY172185 AY172186 AAO65145.1 GAKP01008864 JAC50088.1 GDHF01000973 JAI51341.1 DS235063 EEB11170.1 GDHF01030082 GDHF01007181 JAI22232.1 JAI45133.1 DS232215 EDS37744.1 GDIP01066886 JAM36829.1
BAN66697.1 ODYU01004514 SOQ44475.1 NWSH01006605 PCG63342.1 KU754482 ANW09744.1 KU754481 ANW09743.1 AGBW02009802 OWR49892.1 KQ461103 KPJ09423.1 KQ459606 KPI91774.1 KU754486 ANW09748.1 KU754485 ANW09747.1 GBYB01010845 JAG80612.1 GEMB01002843 JAS00356.1 GECU01031403 GECU01013715 JAS76303.1 JAS93991.1 JTDY01005474 KOB66961.1 GEDC01002338 JAS34960.1 NEVH01003510 PNF40449.1 PNF40448.1 PNF40447.1 GECZ01010984 JAS58785.1 LC368081 BBC77940.1 KQ414736 KOC62134.1 GEBQ01009425 JAT30552.1 KQ434948 KZC12418.1 GBHO01029779 JAG13825.1 GDHC01007506 JAQ11123.1 GBHO01016420 JAG27184.1 GBHO01016419 JAG27185.1 GBRD01008091 GDHC01011007 JAG57730.1 JAQ07622.1 KQ435803 KOX73112.1 GDHC01017351 GDHC01005996 JAQ01278.1 JAQ12633.1 GBRD01005302 JAG60519.1 GBHO01020554 GBRD01005165 JAG23050.1 JAG60656.1 MH105807 AWC08637.1 GBRD01005303 JAG60518.1 GFXV01007876 MBW19681.1 KK852458 KDR23584.1 GGMR01015576 MBY28195.1 GDHF01019143 JAI33171.1 GGMR01004833 MBY17452.1 GBRD01005164 GDHC01016572 JAG60657.1 JAQ02057.1 GDHF01006670 JAI45644.1 GGMR01003601 MBY16220.1 ABLF02038190 UFQS01001196 UFQT01001196 SSX09640.1 SSX29436.1 GAKP01011111 JAC47841.1 ABLF02024643 KY849648 ATY74528.1 GBXI01012586 JAD01706.1 CH940648 EDW62033.1 GECU01009884 JAS97822.1 GEBQ01031928 JAT08049.1 ACPB03020694 GGMS01017627 MBY86830.1 ABLF02024865 GGMS01016781 MBY85984.1 GAMC01010859 JAB95696.1 GECZ01019500 JAS50269.1 GEBQ01021268 JAT18709.1 CP012524 ALC41689.1 AJWK01019808 JRES01000997 KNC26260.1 CH902619 EDV36648.1 IACF01003431 LAB69047.1 GFXV01007001 MBW18806.1 CH933808 EDW09473.1 PYGN01000167 PSN52318.1 CH964282 EDW85642.1 CCAG010001084 KY849652 ATY74530.1 AY172185 AY172186 AAO65145.1 GAKP01008864 JAC50088.1 GDHF01000973 JAI51341.1 DS235063 EEB11170.1 GDHF01030082 GDHF01007181 JAI22232.1 JAI45133.1 DS232215 EDS37744.1 GDIP01066886 JAM36829.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000235965 UP000005203 UP000053825 UP000079169 UP000076502 UP000053105 UP000027135 UP000007819 UP000008792 UP000015103 UP000092553 UP000092461 UP000037069 UP000007801 UP000095301 UP000009192 UP000245037 UP000007798 UP000095300 UP000092445 UP000092444 UP000078200 UP000009046 UP000002320
UP000235965 UP000005203 UP000053825 UP000079169 UP000076502 UP000053105 UP000027135 UP000007819 UP000008792 UP000015103 UP000092553 UP000092461 UP000037069 UP000007801 UP000095301 UP000009192 UP000245037 UP000007798 UP000095300 UP000092445 UP000092444 UP000078200 UP000009046 UP000002320
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9IZ95
S6CBY2
S6BKX3
S6BR80
A0A2H1VUG9
A0A2A4IVY0
+ More
A0A1C8V608 A0A1C8V604 A0A212F822 A0A194QV63 A0A194PGE3 A0A1C8V610 A0A1C8V606 A0A0C9RQR3 A0A161MGQ8 A0A1B6HNQ6 A0A0L7KV61 A0A1B6EAM8 A0A2J7RHZ8 A0A2J7RHY9 A0A2J7RHY7 A0A1B6G8L1 A0A088AJS3 A0A2Z5ZAT2 A0A0L7QU96 A0A3Q0J7W2 A0A3Q0J7Y4 A0A1B6M3P2 A0A154PMA4 A0A3Q0IZ92 A0A0A9X9P7 A0A146LTS6 A0A0A9Y4T5 A0A0A9YCE5 A0A0K8SXT9 A0A0N0BFB7 A0A146KYL8 A0A0K8T4R4 A0A0A9Y0I9 A0A2S0X9Z5 A0A0K8T4T1 A0A2H8U0W7 A0A067RUR8 A0A2S2PFL4 A0A0K8V2T9 A0A2S2NKJ3 A0A0K8T554 A0A0K8W3H6 A0A2S2NGC5 J9JZD3 A0A336KXP5 A0A034VX06 J9M761 A0A2H4TET2 A0A1S3DS16 A0A0A1WSU4 B4LMD8 A0A1B6JFE2 A0A1B6K9B6 T1HEX6 A0A2S2RAY9 X1WJG5 A0A2S2R7P5 W8BR39 J9K443 A0A1B6FJA2 A0A1B6L4R2 A0A0M3QV28 A0A1S4G5F6 A0A1B0CN52 A0A0L0C1S9 B3MFG3 A0A2P2I4T6 A0A1I8NA44 A0A2H8TX38 B4KSG8 A0A2P8Z745 B4NMQ3 A0A1I8QE22 A0A1B0ABN1 A0A1B0FJ32 A0A1A9VCA6 A0A2H4TES7 Q867Z8 A0A034W8J1 A0A1I8MRT6 A0A0K8WJK6 E0VCR4 A0A0K8U6H0 B0WZR9 A0A0P5XY92
A0A1C8V608 A0A1C8V604 A0A212F822 A0A194QV63 A0A194PGE3 A0A1C8V610 A0A1C8V606 A0A0C9RQR3 A0A161MGQ8 A0A1B6HNQ6 A0A0L7KV61 A0A1B6EAM8 A0A2J7RHZ8 A0A2J7RHY9 A0A2J7RHY7 A0A1B6G8L1 A0A088AJS3 A0A2Z5ZAT2 A0A0L7QU96 A0A3Q0J7W2 A0A3Q0J7Y4 A0A1B6M3P2 A0A154PMA4 A0A3Q0IZ92 A0A0A9X9P7 A0A146LTS6 A0A0A9Y4T5 A0A0A9YCE5 A0A0K8SXT9 A0A0N0BFB7 A0A146KYL8 A0A0K8T4R4 A0A0A9Y0I9 A0A2S0X9Z5 A0A0K8T4T1 A0A2H8U0W7 A0A067RUR8 A0A2S2PFL4 A0A0K8V2T9 A0A2S2NKJ3 A0A0K8T554 A0A0K8W3H6 A0A2S2NGC5 J9JZD3 A0A336KXP5 A0A034VX06 J9M761 A0A2H4TET2 A0A1S3DS16 A0A0A1WSU4 B4LMD8 A0A1B6JFE2 A0A1B6K9B6 T1HEX6 A0A2S2RAY9 X1WJG5 A0A2S2R7P5 W8BR39 J9K443 A0A1B6FJA2 A0A1B6L4R2 A0A0M3QV28 A0A1S4G5F6 A0A1B0CN52 A0A0L0C1S9 B3MFG3 A0A2P2I4T6 A0A1I8NA44 A0A2H8TX38 B4KSG8 A0A2P8Z745 B4NMQ3 A0A1I8QE22 A0A1B0ABN1 A0A1B0FJ32 A0A1A9VCA6 A0A2H4TES7 Q867Z8 A0A034W8J1 A0A1I8MRT6 A0A0K8WJK6 E0VCR4 A0A0K8U6H0 B0WZR9 A0A0P5XY92
PDB
6HCO
E-value=2.29616e-06,
Score=117
Ontologies
GO
Topology
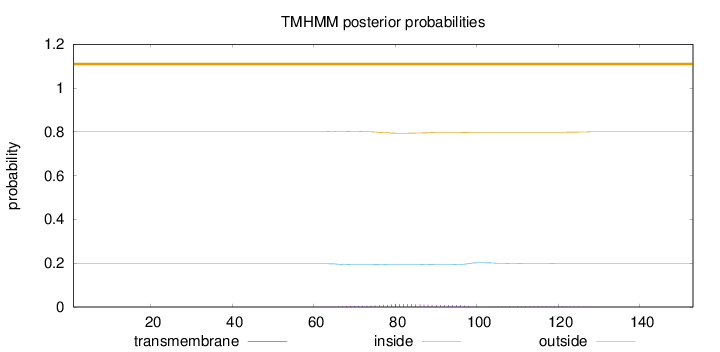
Length:
153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.33967
Exp number, first 60 AAs:
0.00027
Total prob of N-in:
0.19783
outside
1 - 153
Population Genetic Test Statistics
Pi
231.368464
Theta
177.104357
Tajima's D
1.902207
CLR
0.317705
CSRT
0.866606669666517
Interpretation
Uncertain
