Gene
KWMTBOMO02480 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002575
Annotation
PREDICTED:_probable_peroxisomal_acyl-coenzyme_A_oxidase_1_[Amyelois_transitella]
Full name
Acyl-coenzyme A oxidase
+ More
Transaldolase
Transaldolase
Location in the cell
Cytoplasmic Reliability : 2.269
Sequence
CDS
ATGAGCAGCGTGAACCCTGATCTGGTCAAGGAGAGACAGAAATGTACCTTTGACGTACAGGAACTGGTGCATTTCATTGACAGAGGAGAGGAACACACCTTGGAGAGGAAAGAGTTGGAGGATTTGGTGCTTAGTGTCAAAGAGTTGAAGGATGAAACTCCCGAAGAATATTTAAGTCACAAAGAACGCTACGAGAACGCAATCCGAAAGGCCAGTTTATTGTTTAAAATTCTCCAGAAATATTTCGAGACGCACAACTTATTGGACATATTCAGTCCAACAAACAAGTACCGGGTGCAGTTCGGAGTCCTCAAGGATGTGTCACCGTTTTTGGTTCACATGGGCATGTTCCTGCCGACGCTCTTGCATCAAACCACTCCCGAACAAGCAGCCGAGTGGGTCCCTAAAGCGCAGAGCATGCAGATGATCGGCACTTATGCACAGACGGAGCTGGGTCACGGCACCTTCCTGAGAGGCTTGGAGACGACAGCAACTTACGACGCGAAAGCCGAAGAGTTTATACTGAACACTCCAACGCTGACTTCATTCAAATGGTGGCCCGGTGGATTGGCCCACACCGCCAACCACTGCGTTGTGGTAGCCCAACTATACACAAAAGGAAAGTGCTATGGTGTTCATCCGTTCCTCGTGCAAATTCGGGACTTGGAAACGCACATGCCGATGCCTGGTATCAAAATTGGCGAAATTGGATCGAAGCTTGGTTACAACACGGTCAACAATGGTTTCCTTGGCTTCAACAAAGTTAGAATACCGAGGACTAATCTTCTGATGAAGAATGCTCAAGTTCTGAAGGACGGGACTTATGTAAAAGCGAAAAGCGAGAAGTTGACTTATGGCACGATGGTATTGATACGCGTTCTGATTGTCACTGATATGGCGTACGAATTGGCCCGAGCGTCCACCGTCGCTGTCAGGTACTCGGTCGTCCGGCACCAGTCGCAACCGAAGCCGGGACAACCGGAGCCACAGATTATTGAATACGTGACTCAACAACATAAGTTGTTCATTGCTGTCGCTACGAGTCACGTTTACCGAATCACTGGGCACTGGCTCTGGGATACTTACTATAAGGTCACCAGCGATTTGGATAAAGGGAACCTTGAACAGTTGCCTGAACTCCACGCTGTCTCCTGTTGTCTCAAGGCGGTCTGCTCTCGAGACGCCTCAATCCACATCGACGATTGCAGGCTGGCTTGCGGTGGACACGGATACATGAAGTCTTCAAGCTTCCCTTTTATTCAGGCCATCGTAACGGCTAGTGTGACTTACGAAGGAGAATACACGGTTATGATGCTGCAGACTGCAAGATATCTTATGAAGACTTTAAAGCAAGCATTGTCTGGTAAAGCGATAACATCAACTGTGGCCTATCTGTTGGACTTTGACAAGAACAGTAATCGTAATTGGCAGAATACTCCCGAAGGCATCGTTATAGGGTTCCAGGCAATCGCAGCAGGATTAACAAAGGCAGCTTACGATGAAATAGAGAAACACGAAAAGACTGGATTAGATTACGAAGACTCTTGGAATCAGGCTTCCGTGCAACTTGTGAGAGCTAGTGAGGCACATTGCCGGGTGATTATCTGCGAGAAATCTTGGCTTGAAGCGCAGCGACTCACGAAATCGCTCTCCCCTGCTTTGTCGAAGGTGTTGCTGGAATTGGTTGAATTGTACATTATTTATTGGGCGTTGGAGAAAAAGGGAGACTTTCTAGTGTATACGAAAATGCAAAAGGATGACGTTATGCAACTACAGCAGAGATATGAAGAGTTGCTGGCAGTAATTCGACCTAACGCAGTCGGTTTAGTGGACGGCTTCGACATCAGAGACGAGATTCTGAACTCCACACTGGGGGCGTACGACGGGCGCGTGTACGAGCGCCTGATGGAGGAGGCGCTGAAGAGTCCTCTCAACGCGGAGCCCGTTAACACATCCTTCCACCAGTACCTCAAACCTTTCATGACTGCGAAACTGTAA
Protein
MSSVNPDLVKERQKCTFDVQELVHFIDRGEEHTLERKELEDLVLSVKELKDETPEEYLSHKERYENAIRKASLLFKILQKYFETHNLLDIFSPTNKYRVQFGVLKDVSPFLVHMGMFLPTLLHQTTPEQAAEWVPKAQSMQMIGTYAQTELGHGTFLRGLETTATYDAKAEEFILNTPTLTSFKWWPGGLAHTANHCVVVAQLYTKGKCYGVHPFLVQIRDLETHMPMPGIKIGEIGSKLGYNTVNNGFLGFNKVRIPRTNLLMKNAQVLKDGTYVKAKSEKLTYGTMVLIRVLIVTDMAYELARASTVAVRYSVVRHQSQPKPGQPEPQIIEYVTQQHKLFIAVATSHVYRITGHWLWDTYYKVTSDLDKGNLEQLPELHAVSCCLKAVCSRDASIHIDDCRLACGGHGYMKSSSFPFIQAIVTASVTYEGEYTVMMLQTARYLMKTLKQALSGKAITSTVAYLLDFDKNSNRNWQNTPEGIVIGFQAIAAGLTKAAYDEIEKHEKTGLDYEDSWNQASVQLVRASEAHCRVIICEKSWLEAQRLTKSLSPALSKVLLELVELYIIYWALEKKGDFLVYTKMQKDDVMQLQQRYEELLAVIRPNAVGLVDGFDIRDEILNSTLGAYDGRVYERLMEEALKSPLNAEPVNTSFHQYLKPFMTAKL
Summary
Description
Transaldolase is important for the balance of metabolites in the pentose-phosphate pathway.
Catalytic Activity
D-glyceraldehyde 3-phosphate + D-sedoheptulose 7-phosphate = beta-D-fructose 6-phosphate + D-erythrose 4-phosphate
Similarity
Belongs to the acyl-CoA oxidase family.
Feature
chain Acyl-coenzyme A oxidase
Uniprot
H9IZ90
H9IZ89
A0A2A4IZ14
A0A2H1WT00
A0A194Q5G5
A0A2W1BIG1
+ More
A0A212EHT3 A0A194Q4H5 A0A068FKD3 A0A2A4IZR4 A0A2A4IYK0 A0A2W1BMG8 A0A212EHS3 A0A194Q3Y6 H9IZN1 A0A2J7PF41 A0A2H1WT19 H9IZ91 A0A194Q3Y2 A0A067RUS8 A0A2A4IYW9 A0A194Q9Z7 A0A194QVT5 A0A1L8DZ22 E2C709 A0A1L8DZK4 A0A026WLZ8 F4WDF3 E2A9R1 A0A212EHS8 A0A1B6LZ34 H9IZ88 A0A195CBX2 A0A068FKE0 A0A154PF92 A0A1B6GE28 A0A151X0N8 B0WFZ7 A0A2A4J068 A0A1B6DLI0 A0A1B6IC96 A0A232EXB3 A0A2A3EGT9 A0A222YT14 K7JBZ6 D6W793 E0W4E2 A0A1B0CLN4 A0A0C9QVX1 A0A1Q3FGB3 A0A0L7QU29 A0A1Q3FG32 A0A084WHR2 A0A1Y1M345 A0A182FVD2 A0A182M732 A0A088ARZ1 A0A2M4BG86 A0A2M4BGH0 W5JFQ1 A0A182RVB5 A0A182YL67 A0A2M4AAA7 Q16YD0 A0A224X8U0 A0A023FBQ9 A0A340TBF5 A0A2H1WMZ1 A0A2M4AAY4 A0A2M4AAK2 A0A2M4BGC9 A0A2M4BGD3 A0A023EVR9 A0A3F2YW21 A0A2M4BGE4 A0A2M3ZHJ0 A0A336LRR1 A0A182ML30 A0A151J7S0 A0A2M3ZHH0 Q7PZ92 E9IZQ4 A0A1Y9J0Y0 A0A182UN38 A0A0V0G8B7 A0A340TBE9 A0A3F2YW22 T1HSM5 A0A1Y9J094 A0A182QLI0 A0A1Y9GLQ2 U5EW79 A0A182UM20 A0A0A9YPZ1 A0A1Y9GKJ9 A0A182IZK6 W5JFW3 A0A0L7LIQ5 A0A3F2YUR4 J9K7D8
A0A212EHT3 A0A194Q4H5 A0A068FKD3 A0A2A4IZR4 A0A2A4IYK0 A0A2W1BMG8 A0A212EHS3 A0A194Q3Y6 H9IZN1 A0A2J7PF41 A0A2H1WT19 H9IZ91 A0A194Q3Y2 A0A067RUS8 A0A2A4IYW9 A0A194Q9Z7 A0A194QVT5 A0A1L8DZ22 E2C709 A0A1L8DZK4 A0A026WLZ8 F4WDF3 E2A9R1 A0A212EHS8 A0A1B6LZ34 H9IZ88 A0A195CBX2 A0A068FKE0 A0A154PF92 A0A1B6GE28 A0A151X0N8 B0WFZ7 A0A2A4J068 A0A1B6DLI0 A0A1B6IC96 A0A232EXB3 A0A2A3EGT9 A0A222YT14 K7JBZ6 D6W793 E0W4E2 A0A1B0CLN4 A0A0C9QVX1 A0A1Q3FGB3 A0A0L7QU29 A0A1Q3FG32 A0A084WHR2 A0A1Y1M345 A0A182FVD2 A0A182M732 A0A088ARZ1 A0A2M4BG86 A0A2M4BGH0 W5JFQ1 A0A182RVB5 A0A182YL67 A0A2M4AAA7 Q16YD0 A0A224X8U0 A0A023FBQ9 A0A340TBF5 A0A2H1WMZ1 A0A2M4AAY4 A0A2M4AAK2 A0A2M4BGC9 A0A2M4BGD3 A0A023EVR9 A0A3F2YW21 A0A2M4BGE4 A0A2M3ZHJ0 A0A336LRR1 A0A182ML30 A0A151J7S0 A0A2M3ZHH0 Q7PZ92 E9IZQ4 A0A1Y9J0Y0 A0A182UN38 A0A0V0G8B7 A0A340TBE9 A0A3F2YW22 T1HSM5 A0A1Y9J094 A0A182QLI0 A0A1Y9GLQ2 U5EW79 A0A182UM20 A0A0A9YPZ1 A0A1Y9GKJ9 A0A182IZK6 W5JFW3 A0A0L7LIQ5 A0A3F2YUR4 J9K7D8
EC Number
2.2.1.2
Pubmed
EMBL
BABH01014526
BABH01014527
NWSH01004519
PCG65001.1
ODYU01010838
SOQ56189.1
+ More
KQ459465 KPJ00250.1 KZ150070 PZC74071.1 AGBW02014804 OWR41037.1 KPJ00249.1 KJ622089 AID66679.1 NWSH01004692 PCG64764.1 PCG65017.1 PZC74066.1 OWR41036.1 KPJ00253.1 BABH01014529 BABH01014530 BABH01014531 NEVH01026085 PNF14948.1 SOQ56188.1 BABH01014523 BABH01014524 BABH01014525 KPJ00248.1 KK852458 KDR23594.1 PCG65015.1 KPJ00251.1 KQ461103 KPJ09429.1 GFDF01002363 JAV11721.1 GL453255 EFN76281.1 GFDF01002362 JAV11722.1 KK107152 QOIP01000007 EZA57082.1 RLU20599.1 GL888087 EGI67733.1 GL437918 EFN69863.1 OWR41035.1 GEBQ01011024 JAT28953.1 BABH01014528 KQ978068 KYM97593.1 KJ622094 AID66684.1 KQ434892 KZC10515.1 GECZ01009098 JAS60671.1 KQ982603 KYQ53952.1 DS231921 EDS26532.1 PCG64763.1 GEDC01010765 JAS26533.1 GECU01023161 JAS84545.1 NNAY01001755 OXU23026.1 KZ288266 PBC30221.1 KX650075 ASR73781.1 AAZX01008228 KQ971307 EFA11530.1 DS235886 EEB20498.1 AJWK01017463 GBYB01004782 JAG74549.1 GFDL01008460 JAV26585.1 KQ414736 KOC62130.1 GFDL01008542 JAV26503.1 ATLV01023873 ATLV01023874 ATLV01023875 KE525347 KFB49756.1 GEZM01041563 JAV80282.1 AXCM01002223 GGFJ01002935 MBW52076.1 GGFJ01002932 MBW52073.1 ADMH02001629 ETN61710.1 GGFK01004406 MBW37727.1 CH477518 EAT39640.1 GFTR01007681 JAW08745.1 GBBI01000288 JAC18424.1 ODYU01009780 SOQ54435.1 GGFK01004608 MBW37929.1 GGFK01004495 MBW37816.1 GGFJ01002975 MBW52116.1 GGFJ01002974 MBW52115.1 GAPW01000427 JAC13171.1 GGFJ01002973 MBW52114.1 GGFM01007220 MBW27971.1 UFQS01000063 UFQT01000063 SSW98853.1 SSX19239.1 AXCM01002222 KQ979695 KYN19717.1 GGFM01007212 MBW27963.1 AAAB01008986 EAA00765.4 GL767232 EFZ13917.1 GECL01001862 JAP04262.1 ACPB03009151 AXCN02001109 APCN01002398 GANO01001544 JAB58327.1 GBHO01010451 JAG33153.1 ETN61709.1 JTDY01000978 KOB75219.1 ABLF02036307
KQ459465 KPJ00250.1 KZ150070 PZC74071.1 AGBW02014804 OWR41037.1 KPJ00249.1 KJ622089 AID66679.1 NWSH01004692 PCG64764.1 PCG65017.1 PZC74066.1 OWR41036.1 KPJ00253.1 BABH01014529 BABH01014530 BABH01014531 NEVH01026085 PNF14948.1 SOQ56188.1 BABH01014523 BABH01014524 BABH01014525 KPJ00248.1 KK852458 KDR23594.1 PCG65015.1 KPJ00251.1 KQ461103 KPJ09429.1 GFDF01002363 JAV11721.1 GL453255 EFN76281.1 GFDF01002362 JAV11722.1 KK107152 QOIP01000007 EZA57082.1 RLU20599.1 GL888087 EGI67733.1 GL437918 EFN69863.1 OWR41035.1 GEBQ01011024 JAT28953.1 BABH01014528 KQ978068 KYM97593.1 KJ622094 AID66684.1 KQ434892 KZC10515.1 GECZ01009098 JAS60671.1 KQ982603 KYQ53952.1 DS231921 EDS26532.1 PCG64763.1 GEDC01010765 JAS26533.1 GECU01023161 JAS84545.1 NNAY01001755 OXU23026.1 KZ288266 PBC30221.1 KX650075 ASR73781.1 AAZX01008228 KQ971307 EFA11530.1 DS235886 EEB20498.1 AJWK01017463 GBYB01004782 JAG74549.1 GFDL01008460 JAV26585.1 KQ414736 KOC62130.1 GFDL01008542 JAV26503.1 ATLV01023873 ATLV01023874 ATLV01023875 KE525347 KFB49756.1 GEZM01041563 JAV80282.1 AXCM01002223 GGFJ01002935 MBW52076.1 GGFJ01002932 MBW52073.1 ADMH02001629 ETN61710.1 GGFK01004406 MBW37727.1 CH477518 EAT39640.1 GFTR01007681 JAW08745.1 GBBI01000288 JAC18424.1 ODYU01009780 SOQ54435.1 GGFK01004608 MBW37929.1 GGFK01004495 MBW37816.1 GGFJ01002975 MBW52116.1 GGFJ01002974 MBW52115.1 GAPW01000427 JAC13171.1 GGFJ01002973 MBW52114.1 GGFM01007220 MBW27971.1 UFQS01000063 UFQT01000063 SSW98853.1 SSX19239.1 AXCM01002222 KQ979695 KYN19717.1 GGFM01007212 MBW27963.1 AAAB01008986 EAA00765.4 GL767232 EFZ13917.1 GECL01001862 JAP04262.1 ACPB03009151 AXCN02001109 APCN01002398 GANO01001544 JAB58327.1 GBHO01010451 JAG33153.1 ETN61709.1 JTDY01000978 KOB75219.1 ABLF02036307
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000235965
UP000027135
+ More
UP000053240 UP000008237 UP000053097 UP000279307 UP000007755 UP000000311 UP000078542 UP000076502 UP000075809 UP000002320 UP000215335 UP000242457 UP000002358 UP000007266 UP000009046 UP000092461 UP000053825 UP000030765 UP000069272 UP000075883 UP000005203 UP000000673 UP000075900 UP000076408 UP000008820 UP000075920 UP000075884 UP000078492 UP000007062 UP000076407 UP000075903 UP000015103 UP000075886 UP000075840 UP000075880 UP000037510 UP000075882 UP000007819
UP000053240 UP000008237 UP000053097 UP000279307 UP000007755 UP000000311 UP000078542 UP000076502 UP000075809 UP000002320 UP000215335 UP000242457 UP000002358 UP000007266 UP000009046 UP000092461 UP000053825 UP000030765 UP000069272 UP000075883 UP000005203 UP000000673 UP000075900 UP000076408 UP000008820 UP000075920 UP000075884 UP000078492 UP000007062 UP000076407 UP000075903 UP000015103 UP000075886 UP000075840 UP000075880 UP000037510 UP000075882 UP000007819
Pfam
Interpro
IPR029320
Acyl-CoA_ox_N
+ More
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR002655 Acyl-CoA_oxidase_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR012258 Acyl-CoA_oxidase
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR018225 Transaldolase_AS
IPR013785 Aldolase_TIM
IPR001585 TAL/FSA
IPR004730 Transaldolase_1
IPR036483 PWI_dom_sf
IPR002483 PWI_dom
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR002655 Acyl-CoA_oxidase_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR012258 Acyl-CoA_oxidase
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR018225 Transaldolase_AS
IPR013785 Aldolase_TIM
IPR001585 TAL/FSA
IPR004730 Transaldolase_1
IPR036483 PWI_dom_sf
IPR002483 PWI_dom
Gene 3D
ProteinModelPortal
H9IZ90
H9IZ89
A0A2A4IZ14
A0A2H1WT00
A0A194Q5G5
A0A2W1BIG1
+ More
A0A212EHT3 A0A194Q4H5 A0A068FKD3 A0A2A4IZR4 A0A2A4IYK0 A0A2W1BMG8 A0A212EHS3 A0A194Q3Y6 H9IZN1 A0A2J7PF41 A0A2H1WT19 H9IZ91 A0A194Q3Y2 A0A067RUS8 A0A2A4IYW9 A0A194Q9Z7 A0A194QVT5 A0A1L8DZ22 E2C709 A0A1L8DZK4 A0A026WLZ8 F4WDF3 E2A9R1 A0A212EHS8 A0A1B6LZ34 H9IZ88 A0A195CBX2 A0A068FKE0 A0A154PF92 A0A1B6GE28 A0A151X0N8 B0WFZ7 A0A2A4J068 A0A1B6DLI0 A0A1B6IC96 A0A232EXB3 A0A2A3EGT9 A0A222YT14 K7JBZ6 D6W793 E0W4E2 A0A1B0CLN4 A0A0C9QVX1 A0A1Q3FGB3 A0A0L7QU29 A0A1Q3FG32 A0A084WHR2 A0A1Y1M345 A0A182FVD2 A0A182M732 A0A088ARZ1 A0A2M4BG86 A0A2M4BGH0 W5JFQ1 A0A182RVB5 A0A182YL67 A0A2M4AAA7 Q16YD0 A0A224X8U0 A0A023FBQ9 A0A340TBF5 A0A2H1WMZ1 A0A2M4AAY4 A0A2M4AAK2 A0A2M4BGC9 A0A2M4BGD3 A0A023EVR9 A0A3F2YW21 A0A2M4BGE4 A0A2M3ZHJ0 A0A336LRR1 A0A182ML30 A0A151J7S0 A0A2M3ZHH0 Q7PZ92 E9IZQ4 A0A1Y9J0Y0 A0A182UN38 A0A0V0G8B7 A0A340TBE9 A0A3F2YW22 T1HSM5 A0A1Y9J094 A0A182QLI0 A0A1Y9GLQ2 U5EW79 A0A182UM20 A0A0A9YPZ1 A0A1Y9GKJ9 A0A182IZK6 W5JFW3 A0A0L7LIQ5 A0A3F2YUR4 J9K7D8
A0A212EHT3 A0A194Q4H5 A0A068FKD3 A0A2A4IZR4 A0A2A4IYK0 A0A2W1BMG8 A0A212EHS3 A0A194Q3Y6 H9IZN1 A0A2J7PF41 A0A2H1WT19 H9IZ91 A0A194Q3Y2 A0A067RUS8 A0A2A4IYW9 A0A194Q9Z7 A0A194QVT5 A0A1L8DZ22 E2C709 A0A1L8DZK4 A0A026WLZ8 F4WDF3 E2A9R1 A0A212EHS8 A0A1B6LZ34 H9IZ88 A0A195CBX2 A0A068FKE0 A0A154PF92 A0A1B6GE28 A0A151X0N8 B0WFZ7 A0A2A4J068 A0A1B6DLI0 A0A1B6IC96 A0A232EXB3 A0A2A3EGT9 A0A222YT14 K7JBZ6 D6W793 E0W4E2 A0A1B0CLN4 A0A0C9QVX1 A0A1Q3FGB3 A0A0L7QU29 A0A1Q3FG32 A0A084WHR2 A0A1Y1M345 A0A182FVD2 A0A182M732 A0A088ARZ1 A0A2M4BG86 A0A2M4BGH0 W5JFQ1 A0A182RVB5 A0A182YL67 A0A2M4AAA7 Q16YD0 A0A224X8U0 A0A023FBQ9 A0A340TBF5 A0A2H1WMZ1 A0A2M4AAY4 A0A2M4AAK2 A0A2M4BGC9 A0A2M4BGD3 A0A023EVR9 A0A3F2YW21 A0A2M4BGE4 A0A2M3ZHJ0 A0A336LRR1 A0A182ML30 A0A151J7S0 A0A2M3ZHH0 Q7PZ92 E9IZQ4 A0A1Y9J0Y0 A0A182UN38 A0A0V0G8B7 A0A340TBE9 A0A3F2YW22 T1HSM5 A0A1Y9J094 A0A182QLI0 A0A1Y9GLQ2 U5EW79 A0A182UM20 A0A0A9YPZ1 A0A1Y9GKJ9 A0A182IZK6 W5JFW3 A0A0L7LIQ5 A0A3F2YUR4 J9K7D8
PDB
2DDH
E-value=5.17084e-166,
Score=1502
Ontologies
PATHWAY
00071
Fatty acid degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01040 Biosynthesis of unsaturated fatty acids - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01040 Biosynthesis of unsaturated fatty acids - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
GO
Topology
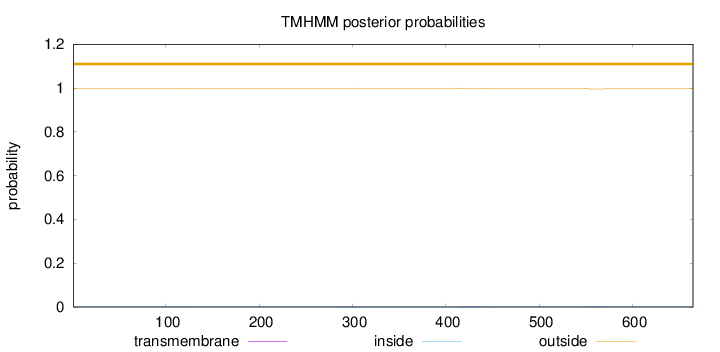
Length:
665
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11056
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00146
outside
1 - 665
Population Genetic Test Statistics
Pi
290.926835
Theta
178.053438
Tajima's D
1.845012
CLR
0.346951
CSRT
0.856957152142393
Interpretation
Uncertain
