Pre Gene Modal
BGIBMGA002719
Annotation
PREDICTED:_translation_initiation_factor_eIF-2B_subunit_delta_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.841 Nuclear Reliability : 1.941
Sequence
CDS
ATGTCTTCTAATGAAATCCCACCAAAGTCTAGAGATGAAATTAAAGCTGCTCGACAGGCAAAAAAAGCTGCCAAACTAAAAGTAAACAACAAGACCACTGAAAATCCTCAACTGACAAAAGAAATAGAGAATCTTAATGAAAACAACATAAATTCTAAACCTAAAGTAGAAAGCACAGATGAAGTAGACAAATCTGTTGTAATAAATCCTAATGCTGTAGATATAAGTACTAGTAGAGAACAGATCAAAGCAGATAGAGCTGCCAAGAAATCTGCCAAACAGTTAAAGAAAAAAGGAGCTGCCGATGGTGAAAGTAATGTGCAAAGCTCTGTAAAAAAAGAACTTAAAGAAGATGATTCTGTGAAACAAAATGAAGATATGACTGTAAAAGATGTTGTTGAAACTTTAAAAAACATAGTTAGTGTTGCCAAGGAGGTTAATGAAATTACGACAAAAGTAAGTGCCTTGGATTTGGGAGGTAAAAAGCAAGAAGAGACAGGCAAGAGTAAAGCAGAACTTCGGGCTGAACGACGCGCCAAACAGGAGGCTCAGAGAGCTGCCAAACAAGCTGCCGAAAAAAAAGTAGTCAAGCCTAAACTTGAAGCTCCTACAGTAAATGTGGATGTTCCGAAGAAAGAGGAAAAAGTTATTAAATCAAAAACCTTGGAGAAGTCGAAAACAAAGTTGCAGCTCGCACAGCGCTTCAACTGGTTTCAGCATCTTTACACTGAGTTTGACAAGAATCCTTTGAAGAAGATACCCATTAATTCCAACCTACACCCCGCCATCGTGAAACTTGGAGTCCAGTTGTCGTCGCGAGTGGTGAAGGGATCGAACGCCAGATGCATCGCTCTGCTGGACGCCCTCAACAAAATGGTGAGGGACTACGTTCTGCCAGCGAAGACTGAGTTCTCCCGCGGCCTGGAGTCGCACCTGGCCTCCAGCCTGGAGTTCCTGTGGACGACGCGCGAGCCCTGCGCCTCGCAGATCAACGCCGTCAAGTTCTTCCGGCACCACCTCGCGCAGCTACCCAACAACGTCGACGAGTTCGATGCCAAAAAAACCCTTCAAGAAGAGATCGATCGCTACATCAGAGAACAGATCGAAATGGCGGGAGAAGCGATCAGCATCGCCGTTAGAAACAAGATCTCCAATGGCGACACTATACTCACTTATGGATGCTCGTCGCTGATCGAGCAGATACTGTGCGAGGCGTGGGAGGCGGGGCTGCAGTTCCGCACGCTGGTGGTGGGCTCGCGCGTGTCCCCCGACGCCGCGGAGATGCTGCGGCGGCTGTGCGCGCGCGGCCTGCCCTGCACCTACGTAGACTTCACCGCCGTCAACTATGTCATGAAGACCGTGAGCAAGGTGGTGCTGGGCGCGGGCGCGCTGCTGGCCAACGGCGCGGTGGTGAGCGCGGCGGGCACGGCGGGCGTGGCGCTGGCGGCGCGCGCTCGCAACCGCCCCGTGCTCGTCGCCTGCCACACGCACAAGTTCATCGACAGCCTCTACACCGACGCCTTCATGCACAACCAGATAGGAGACCCCGACGATCTTCTGGAAAAATCGGATGATAATTCACCACTAAAAGATTGGAGATCGAATTTAAACTTAACACCACTGAACCTGACTTATGACGTCACACCTCCGTGCCTCATCACCGCCGTCGTCACAGAACTTGCCATATTGCCTTGCACCAGCGCTCCCGTTGTACTCAGATTTAAACTCTCCGAATATGGTGTATAA
Protein
MSSNEIPPKSRDEIKAARQAKKAAKLKVNNKTTENPQLTKEIENLNENNINSKPKVESTDEVDKSVVINPNAVDISTSREQIKADRAAKKSAKQLKKKGAADGESNVQSSVKKELKEDDSVKQNEDMTVKDVVETLKNIVSVAKEVNEITTKVSALDLGGKKQEETGKSKAELRAERRAKQEAQRAAKQAAEKKVVKPKLEAPTVNVDVPKKEEKVIKSKTLEKSKTKLQLAQRFNWFQHLYTEFDKNPLKKIPINSNLHPAIVKLGVQLSSRVVKGSNARCIALLDALNKMVRDYVLPAKTEFSRGLESHLASSLEFLWTTREPCASQINAVKFFRHHLAQLPNNVDEFDAKKTLQEEIDRYIREQIEMAGEAISIAVRNKISNGDTILTYGCSSLIEQILCEAWEAGLQFRTLVVGSRVSPDAAEMLRRLCARGLPCTYVDFTAVNYVMKTVSKVVLGAGALLANGAVVSAAGTAGVALAARARNRPVLVACHTHKFIDSLYTDAFMHNQIGDPDDLLEKSDDNSPLKDWRSNLNLTPLNLTYDVTPPCLITAVVTELAILPCTSAPVVLRFKLSEYGV
Summary
Similarity
Belongs to the eIF-2B alpha/beta/delta subunits family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
IPR000649
IF-2B-related
+ More
IPR037171 NagB/RpiA_transferase-like
IPR020568 Ribosomal_S5_D2-typ_fold
IPR000705 Galactokinase
IPR019539 GalKase_gal-bd
IPR019741 Galactokinase_CS
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR036554 GHMP_kinase_C_sf
IPR006203 GHMP_knse_ATP-bd_CS
IPR013750 GHMP_kinase_C_dom
IPR037171 NagB/RpiA_transferase-like
IPR020568 Ribosomal_S5_D2-typ_fold
IPR000705 Galactokinase
IPR019539 GalKase_gal-bd
IPR019741 Galactokinase_CS
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR036554 GHMP_kinase_C_sf
IPR006203 GHMP_knse_ATP-bd_CS
IPR013750 GHMP_kinase_C_dom
Gene 3D
ProteinModelPortal
PDB
6O9Z
E-value=4.36681e-66,
Score=640
Ontologies
GO
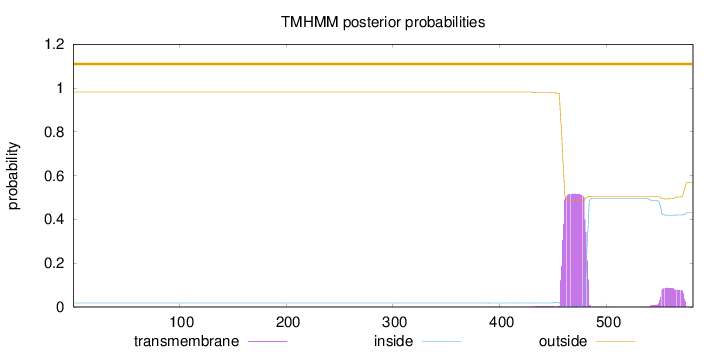
Topology
Length:
581
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.85403
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01793
outside
1 - 581
Population Genetic Test Statistics
Pi
296.200751
Theta
176.357639
Tajima's D
1.638425
CLR
0.534908
CSRT
0.816259187040648
Interpretation
Uncertain
