Gene
KWMTBOMO02470 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002570
Annotation
Dolichyl-diphosphooligosaccharide--protein_glycosyltransferase_subunit_1_[Operophtera_brumata]
Full name
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1
Location in the cell
PlasmaMembrane Reliability : 2.255
Sequence
CDS
ATGAGATTACCGGCAATGGCGAAGTTACAATTTACGCTTTTTTTCGTATTGTTTCTGACGATACAAACCCTTAGTGCAAGCGTGGACACAATATCTAGTGATATAAAGTTAAAAAATGTCGATCGCACAGTAGATATATCGACCCAACTCGTAAAAGTAGTATCAAAAATAGTCTTGGAGAACAGTGGCAAATCGTCAGTAAAACATTTTCTTTTTGCGGTTGAAGAAGCTGCGAAGAATAATCTAGCGTTTATTGGAGCTAAAGACAGCAATAGTAAAGATCTTCGTCTCGCTGAAACCACTGTGAAAGGTTATGACTATGTTAAATTTTGGCGTGTGGAGCTCAAGGAACCCCTAAACGCCGGTGCCACAGTAAATGTTGTTGTCGAAACTGTGTTCACCAAAGCTTTACAACCATATCCGACAGCTATAACTCAGCAAGAAGACCAGTTTGTGAAATATGTTGGCAATTTGTATACATACTCCCCGTACTATACCTCCACACAAAAAACCAGCGTAATTCTAAATACAAAGACTGTTGAATCATTTACCAAGGTGAAGCCTTTCTCACAAGTTGATGGAACAATTATTTATGGACCGTACGCTAATGTTGGGCCTTTCACTGAAAAGGAGCTCAGCATTCACTACAAAAACAATTCACCATTTTTGACTGTTACTCGCGTCGAAAGATTGATAGAAGTATCACATTGGGGTAATATTGCCATTGAAGAGGTAATTGAAATTGAGCATACAGGAGCCAAGCTCAAAGGTCCGTTTTCTAGGTATGACTATCAACAAGATCATCACAGTGGTCCTGCTAGTGTACGTGCCTACAAGACCCTCCTTCCAGCTTCCGCATCAGACGTATACTATAGAGATACGAATGGTAACATCTCCACATCCAATATGAAGGTGAAAAAAGATTCAGTGGAGTTGGATCTCAGACCTAGATATCCTTTGTTTGGAGGCTGGAGAACACATTACACACTTGGTTACAATGTCCCTAGCTATGAGTATTTATATCACTCTGGTAATGAATACTTGCTGAAAATGAGGTCTATTGATCATATTTTTGATGATATGCAAGTGGATGAATTGGTTACAAAGATCATCTTGCCAGAGGGTTCTACAGGGATCAAGTTAAACATACCTTATAGTGTCACAAGATTGCCAGACAGTCTGCACTTCACATACTTAGATACTAAAGGTCGTCCAGTAATCTCGTTTATAAAGAAAAACATTGTGGAGAACCATATCCAAGATTTTCAAATCCGTTATACATTCCCACGTCTGCTGATGTTGCAGGAACCACTTTTAGTTGTTGGCTTCTTATATACATTGTTTTTGTGTGTTATTATTTATGTAAGATTAGATTTTTCAATTCACAAGTCTGAACATAAAGATTAA
Protein
MRLPAMAKLQFTLFFVLFLTIQTLSASVDTISSDIKLKNVDRTVDISTQLVKVVSKIVLENSGKSSVKHFLFAVEEAAKNNLAFIGAKDSNSKDLRLAETTVKGYDYVKFWRVELKEPLNAGATVNVVVETVFTKALQPYPTAITQQEDQFVKYVGNLYTYSPYYTSTQKTSVILNTKTVESFTKVKPFSQVDGTIIYGPYANVGPFTEKELSIHYKNNSPFLTVTRVERLIEVSHWGNIAIEEVIEIEHTGAKLKGPFSRYDYQQDHHSGPASVRAYKTLLPASASDVYYRDTNGNISTSNMKVKKDSVELDLRPRYPLFGGWRTHYTLGYNVPSYEYLYHSGNEYLLKMRSIDHIFDDMQVDELVTKIILPEGSTGIKLNIPYSVTRLPDSLHFTYLDTKGRPVISFIKKNIVENHIQDFQIRYTFPRLLMLQEPLLVVGFLYTLFLCVIIYVRLDFSIHKSEHKD
Summary
Description
Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity.
Subunit
Component of the oligosaccharyltransferase (OST) complex.
Similarity
Belongs to the OST1 family.
Feature
chain Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1
Uniprot
H9IZ85
A0A2A4J0F2
A0A2H1WUP9
M4M4L9
A0A2W1BNG9
A0A194QWV6
+ More
A0A0L7L025 S4NS98 A0A212ERF2 A0A1B6MNY6 A0A1B0DGC7 A0A0A9YE32 Q1HQN9 Q16K94 A0A182GHD5 A0A023EUS8 A0A0V0G2R6 A0A1B6I7Z6 A0A067RA37 A0A1L8E285 A0A182TKM9 A0A182JHT3 A0A182JWB6 U5EUI5 A0A182T0P0 A0A182URK0 A0A224X9L2 R4FP91 A0A182QA06 A0A182XB20 A0A2R7WX54 A0A182I0L6 A0A182KJN1 Q7Q0T4 A0A171AF01 A0A084VQ08 A0A182N8M8 A0A1Q3FC91 A0A182P2B9 A0A336LXT2 B0W726 A0A182R7S3 A0A182WA74 A0A2M4A0S2 A0A2M4A0R1 A0A2M4A135 T1JMK6 A0A182MMF3 A0A182YRQ0 A0A1J1IUI7 A0A1B6D3Z8 A0A2M3Z058 W5JEA2 A0A2M4BME7 A0A310SPA5 K7J1Y5 A0A2T7NTC6 A0A0L7RAC9 A0A0J7KVM3 A0A210PHZ0 E9IHB6 A0A195EWC4 A0A151ICP8 A0A154PQV6 F4WVA4 A0A0N0BE65 A0A226DDL0 R7TCG8 A0A151IWU4 A0A1Y1MSJ9 A0A2A3ERQ0 A0A195BRY9 A0A158NNV6 A0A0B6ZBE4 A0A1S3GYD0 E2A8B3 A0A2P8Z9E7 K1QWZ6 I3KWY2 A0A1A8IPJ4 A0A1A8FE16 A0A1A8S2N5 A0A1A8M988 A0A1A8DRZ5 A0A1A8A5Q3 D6W7G4 A0A1A7XPU8 A0A3B4H0D4 A0A3P9PWW6 A0A026WQQ0 V3Z1F4 A0A1E1X980 A0A2I4BC18 A0A023FPR2 A0A3B4H0E4 A0A3P9BXA8 A0A0S7HUS8 A0A0C9RYB8 A0A3B3V5G4
A0A0L7L025 S4NS98 A0A212ERF2 A0A1B6MNY6 A0A1B0DGC7 A0A0A9YE32 Q1HQN9 Q16K94 A0A182GHD5 A0A023EUS8 A0A0V0G2R6 A0A1B6I7Z6 A0A067RA37 A0A1L8E285 A0A182TKM9 A0A182JHT3 A0A182JWB6 U5EUI5 A0A182T0P0 A0A182URK0 A0A224X9L2 R4FP91 A0A182QA06 A0A182XB20 A0A2R7WX54 A0A182I0L6 A0A182KJN1 Q7Q0T4 A0A171AF01 A0A084VQ08 A0A182N8M8 A0A1Q3FC91 A0A182P2B9 A0A336LXT2 B0W726 A0A182R7S3 A0A182WA74 A0A2M4A0S2 A0A2M4A0R1 A0A2M4A135 T1JMK6 A0A182MMF3 A0A182YRQ0 A0A1J1IUI7 A0A1B6D3Z8 A0A2M3Z058 W5JEA2 A0A2M4BME7 A0A310SPA5 K7J1Y5 A0A2T7NTC6 A0A0L7RAC9 A0A0J7KVM3 A0A210PHZ0 E9IHB6 A0A195EWC4 A0A151ICP8 A0A154PQV6 F4WVA4 A0A0N0BE65 A0A226DDL0 R7TCG8 A0A151IWU4 A0A1Y1MSJ9 A0A2A3ERQ0 A0A195BRY9 A0A158NNV6 A0A0B6ZBE4 A0A1S3GYD0 E2A8B3 A0A2P8Z9E7 K1QWZ6 I3KWY2 A0A1A8IPJ4 A0A1A8FE16 A0A1A8S2N5 A0A1A8M988 A0A1A8DRZ5 A0A1A8A5Q3 D6W7G4 A0A1A7XPU8 A0A3B4H0D4 A0A3P9PWW6 A0A026WQQ0 V3Z1F4 A0A1E1X980 A0A2I4BC18 A0A023FPR2 A0A3B4H0E4 A0A3P9BXA8 A0A0S7HUS8 A0A0C9RYB8 A0A3B3V5G4
Pubmed
19121390
28756777
26354079
26227816
23622113
22118469
+ More
25401762 26823975 17204158 17510324 26483478 24945155 24845553 20966253 12364791 14747013 17210077 24438588 25244985 20920257 23761445 20075255 28812685 21282665 21719571 23254933 28004739 21347285 20798317 29403074 22992520 25186727 18362917 19820115 24508170 30249741 28503490 26131772
25401762 26823975 17204158 17510324 26483478 24945155 24845553 20966253 12364791 14747013 17210077 24438588 25244985 20920257 23761445 20075255 28812685 21282665 21719571 23254933 28004739 21347285 20798317 29403074 22992520 25186727 18362917 19820115 24508170 30249741 28503490 26131772
EMBL
BABH01014536
NWSH01004519
PCG65004.1
ODYU01011208
SOQ56799.1
JQ747499
+ More
AGG36456.1 KZ149951 PZC76599.1 KQ461103 KPJ09440.1 JTDY01004081 KOB68629.1 GAIX01014202 JAA78358.1 AGBW02013039 OWR44055.1 GEBQ01002310 JAT37667.1 AJVK01059514 GBHO01015849 GDHC01015925 JAG27755.1 JAQ02704.1 DQ440405 ABF18438.1 CH477971 EAT34721.1 JXUM01063440 JXUM01063441 KQ562250 KXJ76306.1 GAPW01001374 JAC12224.1 GECL01003761 JAP02363.1 GECU01024648 JAS83058.1 KK852597 KDR20590.1 GFDF01001318 JAV12766.1 GANO01003864 JAB56007.1 GFTR01007379 JAW09047.1 ACPB03003015 GAHY01000999 JAA76511.1 AXCN02001856 KK855831 PTY24031.1 APCN01005219 AAAB01008980 EAA13877.4 GEMB01001151 JAS01997.1 ATLV01015102 KE525002 KFB40052.1 GFDL01009850 JAV25195.1 UFQS01000281 UFQT01000281 SSX02370.1 SSX22745.1 DS231851 EDS37300.1 GGFK01001030 MBW34351.1 GGFK01001020 MBW34341.1 GGFK01001027 MBW34348.1 JH431584 AXCM01000013 CVRI01000059 CRL03236.1 GEDC01016993 JAS20305.1 GGFM01001156 MBW21907.1 ADMH02001634 ETN61673.1 GGFJ01005116 MBW54257.1 KQ762207 OAD56059.1 PZQS01000009 PVD24403.1 KQ414618 KOC67815.1 LBMM01002789 KMQ94361.1 NEDP02076678 OWF36091.1 GL763196 EFZ20044.1 KQ981953 KYN32451.1 KQ978062 KYM97708.1 KQ435007 KZC13648.1 GL888384 EGI61870.1 KQ435845 KOX71198.1 LNIX01000021 OXA43685.1 AMQN01013822 KB310521 ELT91404.1 KQ980843 KYN12295.1 GEZM01023178 JAV88559.1 KZ288193 PBC34184.1 KQ976417 KYM89780.1 ADTU01021806 HACG01018386 CEK65251.1 GL437526 EFN70326.1 PYGN01000138 PSN53126.1 JH817786 EKC26046.1 AERX01008647 HAED01012804 HAEE01006999 SBQ99208.1 HAEB01009837 HAEC01009078 SBQ56364.1 HAEI01011010 SBS12217.1 HAEF01011714 HAEG01001932 SBR52869.1 HADZ01014382 HAEA01007858 SBQ36338.1 HADY01011528 HAEJ01001522 SBP50013.1 KQ971307 EFA11223.1 HADW01018440 HADX01008053 SBP19840.1 KK107128 QOIP01000006 EZA58372.1 RLU21877.1 KB203534 ESO84348.1 GFAC01003612 JAT95576.1 GBBK01001652 JAC22830.1 GBYX01436713 JAO44628.1 GBZX01000009 JAG92731.1
AGG36456.1 KZ149951 PZC76599.1 KQ461103 KPJ09440.1 JTDY01004081 KOB68629.1 GAIX01014202 JAA78358.1 AGBW02013039 OWR44055.1 GEBQ01002310 JAT37667.1 AJVK01059514 GBHO01015849 GDHC01015925 JAG27755.1 JAQ02704.1 DQ440405 ABF18438.1 CH477971 EAT34721.1 JXUM01063440 JXUM01063441 KQ562250 KXJ76306.1 GAPW01001374 JAC12224.1 GECL01003761 JAP02363.1 GECU01024648 JAS83058.1 KK852597 KDR20590.1 GFDF01001318 JAV12766.1 GANO01003864 JAB56007.1 GFTR01007379 JAW09047.1 ACPB03003015 GAHY01000999 JAA76511.1 AXCN02001856 KK855831 PTY24031.1 APCN01005219 AAAB01008980 EAA13877.4 GEMB01001151 JAS01997.1 ATLV01015102 KE525002 KFB40052.1 GFDL01009850 JAV25195.1 UFQS01000281 UFQT01000281 SSX02370.1 SSX22745.1 DS231851 EDS37300.1 GGFK01001030 MBW34351.1 GGFK01001020 MBW34341.1 GGFK01001027 MBW34348.1 JH431584 AXCM01000013 CVRI01000059 CRL03236.1 GEDC01016993 JAS20305.1 GGFM01001156 MBW21907.1 ADMH02001634 ETN61673.1 GGFJ01005116 MBW54257.1 KQ762207 OAD56059.1 PZQS01000009 PVD24403.1 KQ414618 KOC67815.1 LBMM01002789 KMQ94361.1 NEDP02076678 OWF36091.1 GL763196 EFZ20044.1 KQ981953 KYN32451.1 KQ978062 KYM97708.1 KQ435007 KZC13648.1 GL888384 EGI61870.1 KQ435845 KOX71198.1 LNIX01000021 OXA43685.1 AMQN01013822 KB310521 ELT91404.1 KQ980843 KYN12295.1 GEZM01023178 JAV88559.1 KZ288193 PBC34184.1 KQ976417 KYM89780.1 ADTU01021806 HACG01018386 CEK65251.1 GL437526 EFN70326.1 PYGN01000138 PSN53126.1 JH817786 EKC26046.1 AERX01008647 HAED01012804 HAEE01006999 SBQ99208.1 HAEB01009837 HAEC01009078 SBQ56364.1 HAEI01011010 SBS12217.1 HAEF01011714 HAEG01001932 SBR52869.1 HADZ01014382 HAEA01007858 SBQ36338.1 HADY01011528 HAEJ01001522 SBP50013.1 KQ971307 EFA11223.1 HADW01018440 HADX01008053 SBP19840.1 KK107128 QOIP01000006 EZA58372.1 RLU21877.1 KB203534 ESO84348.1 GFAC01003612 JAT95576.1 GBBK01001652 JAC22830.1 GBYX01436713 JAO44628.1 GBZX01000009 JAG92731.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000007151
UP000092462
+ More
UP000008820 UP000069940 UP000249989 UP000027135 UP000075902 UP000075880 UP000075881 UP000075901 UP000075903 UP000015103 UP000075886 UP000076407 UP000075840 UP000075882 UP000007062 UP000030765 UP000075884 UP000075885 UP000002320 UP000075900 UP000075920 UP000075883 UP000076408 UP000183832 UP000000673 UP000002358 UP000245119 UP000053825 UP000036403 UP000242188 UP000078541 UP000078542 UP000076502 UP000007755 UP000053105 UP000198287 UP000014760 UP000078492 UP000242457 UP000078540 UP000005205 UP000085678 UP000000311 UP000245037 UP000005408 UP000005207 UP000007266 UP000261460 UP000242638 UP000053097 UP000279307 UP000030746 UP000192220 UP000265160 UP000261500
UP000008820 UP000069940 UP000249989 UP000027135 UP000075902 UP000075880 UP000075881 UP000075901 UP000075903 UP000015103 UP000075886 UP000076407 UP000075840 UP000075882 UP000007062 UP000030765 UP000075884 UP000075885 UP000002320 UP000075900 UP000075920 UP000075883 UP000076408 UP000183832 UP000000673 UP000002358 UP000245119 UP000053825 UP000036403 UP000242188 UP000078541 UP000078542 UP000076502 UP000007755 UP000053105 UP000198287 UP000014760 UP000078492 UP000242457 UP000078540 UP000005205 UP000085678 UP000000311 UP000245037 UP000005408 UP000005207 UP000007266 UP000261460 UP000242638 UP000053097 UP000279307 UP000030746 UP000192220 UP000265160 UP000261500
ProteinModelPortal
H9IZ85
A0A2A4J0F2
A0A2H1WUP9
M4M4L9
A0A2W1BNG9
A0A194QWV6
+ More
A0A0L7L025 S4NS98 A0A212ERF2 A0A1B6MNY6 A0A1B0DGC7 A0A0A9YE32 Q1HQN9 Q16K94 A0A182GHD5 A0A023EUS8 A0A0V0G2R6 A0A1B6I7Z6 A0A067RA37 A0A1L8E285 A0A182TKM9 A0A182JHT3 A0A182JWB6 U5EUI5 A0A182T0P0 A0A182URK0 A0A224X9L2 R4FP91 A0A182QA06 A0A182XB20 A0A2R7WX54 A0A182I0L6 A0A182KJN1 Q7Q0T4 A0A171AF01 A0A084VQ08 A0A182N8M8 A0A1Q3FC91 A0A182P2B9 A0A336LXT2 B0W726 A0A182R7S3 A0A182WA74 A0A2M4A0S2 A0A2M4A0R1 A0A2M4A135 T1JMK6 A0A182MMF3 A0A182YRQ0 A0A1J1IUI7 A0A1B6D3Z8 A0A2M3Z058 W5JEA2 A0A2M4BME7 A0A310SPA5 K7J1Y5 A0A2T7NTC6 A0A0L7RAC9 A0A0J7KVM3 A0A210PHZ0 E9IHB6 A0A195EWC4 A0A151ICP8 A0A154PQV6 F4WVA4 A0A0N0BE65 A0A226DDL0 R7TCG8 A0A151IWU4 A0A1Y1MSJ9 A0A2A3ERQ0 A0A195BRY9 A0A158NNV6 A0A0B6ZBE4 A0A1S3GYD0 E2A8B3 A0A2P8Z9E7 K1QWZ6 I3KWY2 A0A1A8IPJ4 A0A1A8FE16 A0A1A8S2N5 A0A1A8M988 A0A1A8DRZ5 A0A1A8A5Q3 D6W7G4 A0A1A7XPU8 A0A3B4H0D4 A0A3P9PWW6 A0A026WQQ0 V3Z1F4 A0A1E1X980 A0A2I4BC18 A0A023FPR2 A0A3B4H0E4 A0A3P9BXA8 A0A0S7HUS8 A0A0C9RYB8 A0A3B3V5G4
A0A0L7L025 S4NS98 A0A212ERF2 A0A1B6MNY6 A0A1B0DGC7 A0A0A9YE32 Q1HQN9 Q16K94 A0A182GHD5 A0A023EUS8 A0A0V0G2R6 A0A1B6I7Z6 A0A067RA37 A0A1L8E285 A0A182TKM9 A0A182JHT3 A0A182JWB6 U5EUI5 A0A182T0P0 A0A182URK0 A0A224X9L2 R4FP91 A0A182QA06 A0A182XB20 A0A2R7WX54 A0A182I0L6 A0A182KJN1 Q7Q0T4 A0A171AF01 A0A084VQ08 A0A182N8M8 A0A1Q3FC91 A0A182P2B9 A0A336LXT2 B0W726 A0A182R7S3 A0A182WA74 A0A2M4A0S2 A0A2M4A0R1 A0A2M4A135 T1JMK6 A0A182MMF3 A0A182YRQ0 A0A1J1IUI7 A0A1B6D3Z8 A0A2M3Z058 W5JEA2 A0A2M4BME7 A0A310SPA5 K7J1Y5 A0A2T7NTC6 A0A0L7RAC9 A0A0J7KVM3 A0A210PHZ0 E9IHB6 A0A195EWC4 A0A151ICP8 A0A154PQV6 F4WVA4 A0A0N0BE65 A0A226DDL0 R7TCG8 A0A151IWU4 A0A1Y1MSJ9 A0A2A3ERQ0 A0A195BRY9 A0A158NNV6 A0A0B6ZBE4 A0A1S3GYD0 E2A8B3 A0A2P8Z9E7 K1QWZ6 I3KWY2 A0A1A8IPJ4 A0A1A8FE16 A0A1A8S2N5 A0A1A8M988 A0A1A8DRZ5 A0A1A8A5Q3 D6W7G4 A0A1A7XPU8 A0A3B4H0D4 A0A3P9PWW6 A0A026WQQ0 V3Z1F4 A0A1E1X980 A0A2I4BC18 A0A023FPR2 A0A3B4H0E4 A0A3P9BXA8 A0A0S7HUS8 A0A0C9RYB8 A0A3B3V5G4
PDB
6EZN
E-value=4.76636e-29,
Score=319
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
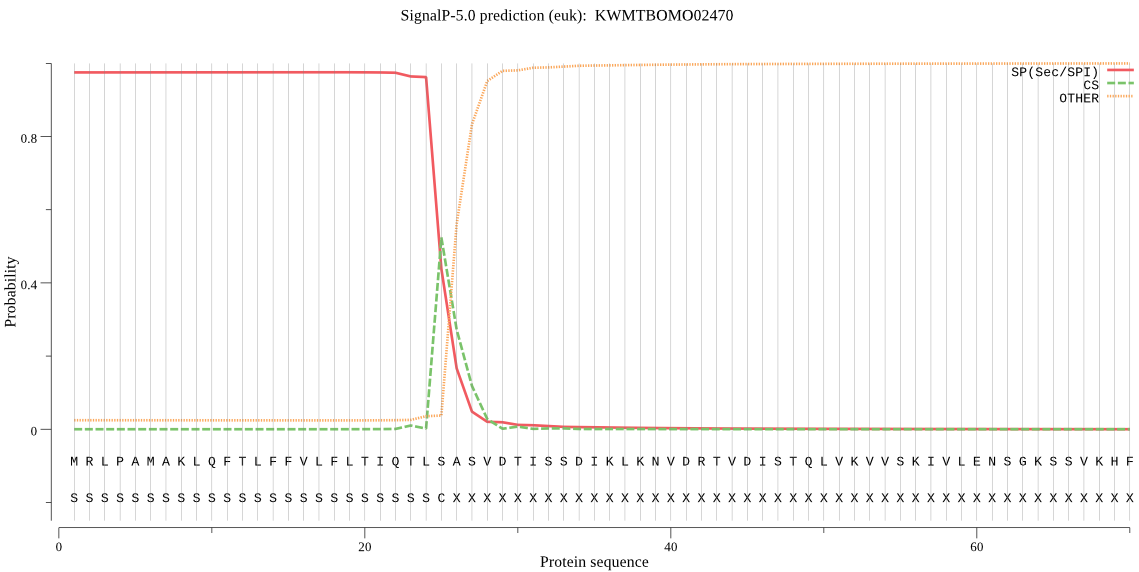
SignalP
Position: 1 - 25,
Likelihood: 0.975462
Length:
468
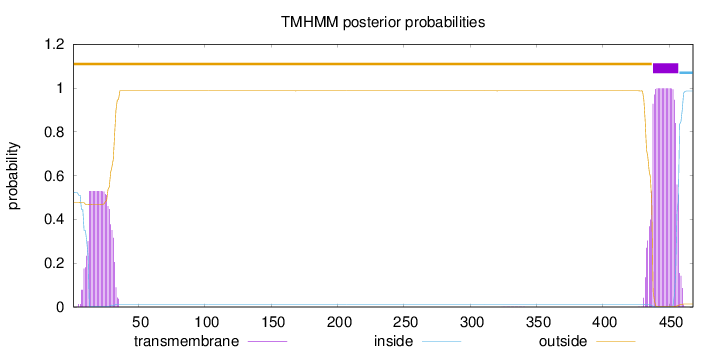
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
31.96234
Exp number, first 60 AAs:
10.78114
Total prob of N-in:
0.52263
POSSIBLE N-term signal
sequence
outside
1 - 437
TMhelix
438 - 457
inside
458 - 468
Population Genetic Test Statistics
Pi
179.823103
Theta
229.244127
Tajima's D
-0.686954
CLR
1011.379785
CSRT
0.20133993300335
Interpretation
Uncertain
