Gene
KWMTBOMO02454
Pre Gene Modal
BGIBMGA002560
Annotation
PREDICTED:_protein_MEF2BNB_homolog_[Amyelois_transitella]
Full name
BLOC-1-related complex subunit 8 homolog
Location in the cell
Nuclear Reliability : 3.472
Sequence
CDS
ATGGCTTTCGTTTACCAAGGAGATGCCGAATTAGAAACTAAAGCAAAGAAGGCTGCTGAAAGAATATCTGAGAATATGCACATCGTAGCCAACGAACCTTCACTAGCACTGTACAGATTACAAGAACATGTAAGGAAGGCCTTGCCTCCTATGGTGGAAAGAAGAGTTGAGGTCACAAAACTACAATATGAATTGCAAGGGAGGTGTTATGATGTTGAATATGCATTAAGTGCTGTAAAATCTATGGATGGTGCTGCAACAAGCTTTAAAAACATACAAGAAATGTTAAAACAGTGCATATTTTTAAAGCAGCAACTAAAATATGAAGAAAGTAGGAGAAATAAAAAAGATTCCAATTCTGTGTACAAACGTCTATCTGCTCATATAGTTTTAGATCTCCCAGACCTGTCCGAGTTCTCCATGGTACGTGAGACCACTAACCGAATCGAGAACATGATGGCTCATGCTAGGGGCTCATCAGCCGAACTCCACAGATCACATACAACTTTACATTGA
Protein
MAFVYQGDAELETKAKKAAERISENMHIVANEPSLALYRLQEHVRKALPPMVERRVEVTKLQYELQGRCYDVEYALSAVKSMDGAATSFKNIQEMLKQCIFLKQQLKYEESRRNKKDSNSVYKRLSAHIVLDLPDLSEFSMVRETTNRIENMMAHARGSSAELHRSHTTLH
Summary
Description
May participate in the coupling of lysosomes to microtubule plus-end-directed kinesin motor.
Similarity
Belongs to the BORCS8 family.
Keywords
Complete proteome
Lysosome
Membrane
Reference proteome
Feature
chain BLOC-1-related complex subunit 8 homolog
Uniprot
H9IZ75
A0A2H1WW93
A0A194QA23
A0A212F2Q1
A0A194QWX4
A0A2A4IWE6
+ More
S4PHR0 A0A0L7L1V4 A0A2J7PMJ1 A0A1L8DCZ7 A0A182WFJ9 A0A1B6HNS5 E2B618 A0A182RK85 A0A1B0CQW8 A0A1B6FX19 A0A1Y1N7K4 A0A182TKI6 A0A182X5Z4 A0A182V191 A0A182KMF6 A0A182YQH7 D6W8B8 A0A0T6AUU6 A0A182P4B7 Q7Q8J2 A0A182KGQ2 A0A1B6LK13 A0A182IBM6 A0A2M4CS84 A0A154PLT5 A0A182F0U2 Q178X2 A0A023EI99 A0A1L8DBR2 A0A2M4C0B6 F4X4S4 A0A151II96 A0A232FKY2 K7J0D9 E2AX29 A0A310SHN1 A0A2A3EV73 V5FV59 A0A182G221 A0A026W583 A0A158NWB3 A0A182GAV6 A0A182NTC6 A0A182ITY3 A0A0L7RBD7 N6U6H3 A0A2M4ATJ2 A0A1B6E3E9 A0A182Q9D2 A0A151HYA6 A0A2M4AW65 A0A336M300 A0A0N0U725 A0A336MCW4 A0A1B0DDX5 A0A1W4XD51 A0A2R7W5U1 A0A182MB38 A0A0C9RFB9 A0A1Q3F5N4 A0A0K8TRB1 B0WUR1 E0VCU2 E9J4Q7 A0A195ET33 A0A023FAU0 A0A069DWG3 R4G3F4 A0A0P4VRT2 A0A1W4XMQ5 T1GZ58 A0A088AJ81 A0A224XMK9 A0A0J7L332 B4JPL5 A0A1A9WEV0 A0A0K8WM55 A0A146LI17 A0A034WBF1 B4LR28 A0A0C9PU33 A0A1B0G0T1 A0A1A9Y1Y4 W8BXC3 A0A0A1WQJ9 A0A0K8SJY2 A0A0K8SII4 A0A1A9UIP7 A0A0Q9X7I6 A0A1A9Z2I7 B4PW74 B3NUY2 Q8IR45 B4R4W3
S4PHR0 A0A0L7L1V4 A0A2J7PMJ1 A0A1L8DCZ7 A0A182WFJ9 A0A1B6HNS5 E2B618 A0A182RK85 A0A1B0CQW8 A0A1B6FX19 A0A1Y1N7K4 A0A182TKI6 A0A182X5Z4 A0A182V191 A0A182KMF6 A0A182YQH7 D6W8B8 A0A0T6AUU6 A0A182P4B7 Q7Q8J2 A0A182KGQ2 A0A1B6LK13 A0A182IBM6 A0A2M4CS84 A0A154PLT5 A0A182F0U2 Q178X2 A0A023EI99 A0A1L8DBR2 A0A2M4C0B6 F4X4S4 A0A151II96 A0A232FKY2 K7J0D9 E2AX29 A0A310SHN1 A0A2A3EV73 V5FV59 A0A182G221 A0A026W583 A0A158NWB3 A0A182GAV6 A0A182NTC6 A0A182ITY3 A0A0L7RBD7 N6U6H3 A0A2M4ATJ2 A0A1B6E3E9 A0A182Q9D2 A0A151HYA6 A0A2M4AW65 A0A336M300 A0A0N0U725 A0A336MCW4 A0A1B0DDX5 A0A1W4XD51 A0A2R7W5U1 A0A182MB38 A0A0C9RFB9 A0A1Q3F5N4 A0A0K8TRB1 B0WUR1 E0VCU2 E9J4Q7 A0A195ET33 A0A023FAU0 A0A069DWG3 R4G3F4 A0A0P4VRT2 A0A1W4XMQ5 T1GZ58 A0A088AJ81 A0A224XMK9 A0A0J7L332 B4JPL5 A0A1A9WEV0 A0A0K8WM55 A0A146LI17 A0A034WBF1 B4LR28 A0A0C9PU33 A0A1B0G0T1 A0A1A9Y1Y4 W8BXC3 A0A0A1WQJ9 A0A0K8SJY2 A0A0K8SII4 A0A1A9UIP7 A0A0Q9X7I6 A0A1A9Z2I7 B4PW74 B3NUY2 Q8IR45 B4R4W3
Pubmed
19121390
26354079
22118469
23622113
26227816
20798317
+ More
28004739 20966253 25244985 18362917 19820115 12364791 14747013 17210077 17510324 24945155 21719571 28648823 20075255 26483478 24508170 30249741 21347285 23537049 26369729 20566863 21282665 25474469 26334808 27129103 17994087 26823975 25348373 18057021 24495485 25830018 17550304 10731132 12537572
28004739 20966253 25244985 18362917 19820115 12364791 14747013 17210077 17510324 24945155 21719571 28648823 20075255 26483478 24508170 30249741 21347285 23537049 26369729 20566863 21282665 25474469 26334808 27129103 17994087 26823975 25348373 18057021 24495485 25830018 17550304 10731132 12537572
EMBL
BABH01014567
ODYU01011494
SOQ57257.1
KQ459465
KPJ00281.1
AGBW02010715
+ More
OWR47991.1 KQ461103 KPJ09460.1 NWSH01006033 PCG63728.1 GAIX01000389 JAA92171.1 JTDY01003501 KOB69452.1 NEVH01023984 PNF17553.1 GFDF01009900 JAV04184.1 GECU01031384 JAS76322.1 GL445900 EFN88879.1 AJWK01024153 GECZ01015021 JAS54748.1 GEZM01010990 JAV93689.1 KQ971307 EFA10886.1 LJIG01022755 KRT78892.1 AAAB01008944 EAA10220.4 GEBQ01015990 GEBQ01008375 JAT23987.1 JAT31602.1 APCN01000711 GGFL01003957 MBW68135.1 KQ434977 KZC12826.1 CH477357 EAT42770.1 GAPW01004957 JAC08641.1 GFDF01010181 JAV03903.1 GGFJ01009619 MBW58760.1 GL888679 EGI58497.1 KQ977505 KYN02271.1 NNAY01000081 OXU31159.1 GL443520 EFN61979.1 KQ768636 OAD53090.1 KZ288185 PBC34971.1 GALX01006936 JAB61530.1 JXUM01006533 KQ560217 KXJ83775.1 KK107405 QOIP01000014 EZA51232.1 RLU14975.1 ADTU01027868 JXUM01051584 KQ561695 KXJ77786.1 KQ414617 KOC68237.1 APGK01047136 KB741077 KB632384 ENN74152.1 ERL94285.1 GGFK01010790 MBW44111.1 GEDC01004854 JAS32444.1 AXCN02000715 KQ976730 KYM76455.1 GGFK01011716 MBW45037.1 UFQT01000328 SSX23293.1 KQ435711 KOX79471.1 UFQT01000943 SSX28145.1 AJVK01005514 KK854239 PTY13575.1 AXCM01005597 GBYB01011807 JAG81574.1 GFDL01012206 JAV22839.1 GDAI01000935 JAI16668.1 DS232108 EDS35047.1 DS235065 EEB11198.1 GL768112 EFZ12207.1 KQ981993 KYN31072.1 GBBI01000674 JAC18038.1 GBGD01003215 JAC85674.1 ACPB03021336 ACPB03021337 GAHY01001726 JAA75784.1 GDKW01001991 JAI54604.1 CAQQ02373773 GFTR01002699 JAW13727.1 LBMM01001019 KMQ97061.1 CH916372 EDV98845.1 GDHF01000096 JAI52218.1 GDHC01010845 JAQ07784.1 GAKP01006051 GAKP01006050 JAC52901.1 CH940649 EDW64567.2 KRF81731.1 KRF81732.1 GBYB01004843 JAG74610.1 CCAG010001884 GAMC01000585 JAC05971.1 GBXI01013003 JAD01289.1 GBRD01012729 JAG53095.1 GBRD01012728 JAG53096.1 CH933807 KRG03193.1 CM000162 EDX01705.1 CH954180 EDV47080.1 AE014298 BT025828 CM000366 EDX17920.1
OWR47991.1 KQ461103 KPJ09460.1 NWSH01006033 PCG63728.1 GAIX01000389 JAA92171.1 JTDY01003501 KOB69452.1 NEVH01023984 PNF17553.1 GFDF01009900 JAV04184.1 GECU01031384 JAS76322.1 GL445900 EFN88879.1 AJWK01024153 GECZ01015021 JAS54748.1 GEZM01010990 JAV93689.1 KQ971307 EFA10886.1 LJIG01022755 KRT78892.1 AAAB01008944 EAA10220.4 GEBQ01015990 GEBQ01008375 JAT23987.1 JAT31602.1 APCN01000711 GGFL01003957 MBW68135.1 KQ434977 KZC12826.1 CH477357 EAT42770.1 GAPW01004957 JAC08641.1 GFDF01010181 JAV03903.1 GGFJ01009619 MBW58760.1 GL888679 EGI58497.1 KQ977505 KYN02271.1 NNAY01000081 OXU31159.1 GL443520 EFN61979.1 KQ768636 OAD53090.1 KZ288185 PBC34971.1 GALX01006936 JAB61530.1 JXUM01006533 KQ560217 KXJ83775.1 KK107405 QOIP01000014 EZA51232.1 RLU14975.1 ADTU01027868 JXUM01051584 KQ561695 KXJ77786.1 KQ414617 KOC68237.1 APGK01047136 KB741077 KB632384 ENN74152.1 ERL94285.1 GGFK01010790 MBW44111.1 GEDC01004854 JAS32444.1 AXCN02000715 KQ976730 KYM76455.1 GGFK01011716 MBW45037.1 UFQT01000328 SSX23293.1 KQ435711 KOX79471.1 UFQT01000943 SSX28145.1 AJVK01005514 KK854239 PTY13575.1 AXCM01005597 GBYB01011807 JAG81574.1 GFDL01012206 JAV22839.1 GDAI01000935 JAI16668.1 DS232108 EDS35047.1 DS235065 EEB11198.1 GL768112 EFZ12207.1 KQ981993 KYN31072.1 GBBI01000674 JAC18038.1 GBGD01003215 JAC85674.1 ACPB03021336 ACPB03021337 GAHY01001726 JAA75784.1 GDKW01001991 JAI54604.1 CAQQ02373773 GFTR01002699 JAW13727.1 LBMM01001019 KMQ97061.1 CH916372 EDV98845.1 GDHF01000096 JAI52218.1 GDHC01010845 JAQ07784.1 GAKP01006051 GAKP01006050 JAC52901.1 CH940649 EDW64567.2 KRF81731.1 KRF81732.1 GBYB01004843 JAG74610.1 CCAG010001884 GAMC01000585 JAC05971.1 GBXI01013003 JAD01289.1 GBRD01012729 JAG53095.1 GBRD01012728 JAG53096.1 CH933807 KRG03193.1 CM000162 EDX01705.1 CH954180 EDV47080.1 AE014298 BT025828 CM000366 EDX17920.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000218220
UP000037510
+ More
UP000235965 UP000075920 UP000008237 UP000075900 UP000092461 UP000075902 UP000076407 UP000075903 UP000075882 UP000076408 UP000007266 UP000075885 UP000007062 UP000075881 UP000075840 UP000076502 UP000069272 UP000008820 UP000007755 UP000078542 UP000215335 UP000002358 UP000000311 UP000242457 UP000069940 UP000249989 UP000053097 UP000279307 UP000005205 UP000075884 UP000075880 UP000053825 UP000019118 UP000030742 UP000075886 UP000078540 UP000053105 UP000092462 UP000192223 UP000075883 UP000002320 UP000009046 UP000078541 UP000015103 UP000015102 UP000005203 UP000036403 UP000001070 UP000091820 UP000008792 UP000092444 UP000092443 UP000078200 UP000009192 UP000092445 UP000002282 UP000008711 UP000000803 UP000000304
UP000235965 UP000075920 UP000008237 UP000075900 UP000092461 UP000075902 UP000076407 UP000075903 UP000075882 UP000076408 UP000007266 UP000075885 UP000007062 UP000075881 UP000075840 UP000076502 UP000069272 UP000008820 UP000007755 UP000078542 UP000215335 UP000002358 UP000000311 UP000242457 UP000069940 UP000249989 UP000053097 UP000279307 UP000005205 UP000075884 UP000075880 UP000053825 UP000019118 UP000030742 UP000075886 UP000078540 UP000053105 UP000092462 UP000192223 UP000075883 UP000002320 UP000009046 UP000078541 UP000015103 UP000015102 UP000005203 UP000036403 UP000001070 UP000091820 UP000008792 UP000092444 UP000092443 UP000078200 UP000009192 UP000092445 UP000002282 UP000008711 UP000000803 UP000000304
Interpro
ProteinModelPortal
H9IZ75
A0A2H1WW93
A0A194QA23
A0A212F2Q1
A0A194QWX4
A0A2A4IWE6
+ More
S4PHR0 A0A0L7L1V4 A0A2J7PMJ1 A0A1L8DCZ7 A0A182WFJ9 A0A1B6HNS5 E2B618 A0A182RK85 A0A1B0CQW8 A0A1B6FX19 A0A1Y1N7K4 A0A182TKI6 A0A182X5Z4 A0A182V191 A0A182KMF6 A0A182YQH7 D6W8B8 A0A0T6AUU6 A0A182P4B7 Q7Q8J2 A0A182KGQ2 A0A1B6LK13 A0A182IBM6 A0A2M4CS84 A0A154PLT5 A0A182F0U2 Q178X2 A0A023EI99 A0A1L8DBR2 A0A2M4C0B6 F4X4S4 A0A151II96 A0A232FKY2 K7J0D9 E2AX29 A0A310SHN1 A0A2A3EV73 V5FV59 A0A182G221 A0A026W583 A0A158NWB3 A0A182GAV6 A0A182NTC6 A0A182ITY3 A0A0L7RBD7 N6U6H3 A0A2M4ATJ2 A0A1B6E3E9 A0A182Q9D2 A0A151HYA6 A0A2M4AW65 A0A336M300 A0A0N0U725 A0A336MCW4 A0A1B0DDX5 A0A1W4XD51 A0A2R7W5U1 A0A182MB38 A0A0C9RFB9 A0A1Q3F5N4 A0A0K8TRB1 B0WUR1 E0VCU2 E9J4Q7 A0A195ET33 A0A023FAU0 A0A069DWG3 R4G3F4 A0A0P4VRT2 A0A1W4XMQ5 T1GZ58 A0A088AJ81 A0A224XMK9 A0A0J7L332 B4JPL5 A0A1A9WEV0 A0A0K8WM55 A0A146LI17 A0A034WBF1 B4LR28 A0A0C9PU33 A0A1B0G0T1 A0A1A9Y1Y4 W8BXC3 A0A0A1WQJ9 A0A0K8SJY2 A0A0K8SII4 A0A1A9UIP7 A0A0Q9X7I6 A0A1A9Z2I7 B4PW74 B3NUY2 Q8IR45 B4R4W3
S4PHR0 A0A0L7L1V4 A0A2J7PMJ1 A0A1L8DCZ7 A0A182WFJ9 A0A1B6HNS5 E2B618 A0A182RK85 A0A1B0CQW8 A0A1B6FX19 A0A1Y1N7K4 A0A182TKI6 A0A182X5Z4 A0A182V191 A0A182KMF6 A0A182YQH7 D6W8B8 A0A0T6AUU6 A0A182P4B7 Q7Q8J2 A0A182KGQ2 A0A1B6LK13 A0A182IBM6 A0A2M4CS84 A0A154PLT5 A0A182F0U2 Q178X2 A0A023EI99 A0A1L8DBR2 A0A2M4C0B6 F4X4S4 A0A151II96 A0A232FKY2 K7J0D9 E2AX29 A0A310SHN1 A0A2A3EV73 V5FV59 A0A182G221 A0A026W583 A0A158NWB3 A0A182GAV6 A0A182NTC6 A0A182ITY3 A0A0L7RBD7 N6U6H3 A0A2M4ATJ2 A0A1B6E3E9 A0A182Q9D2 A0A151HYA6 A0A2M4AW65 A0A336M300 A0A0N0U725 A0A336MCW4 A0A1B0DDX5 A0A1W4XD51 A0A2R7W5U1 A0A182MB38 A0A0C9RFB9 A0A1Q3F5N4 A0A0K8TRB1 B0WUR1 E0VCU2 E9J4Q7 A0A195ET33 A0A023FAU0 A0A069DWG3 R4G3F4 A0A0P4VRT2 A0A1W4XMQ5 T1GZ58 A0A088AJ81 A0A224XMK9 A0A0J7L332 B4JPL5 A0A1A9WEV0 A0A0K8WM55 A0A146LI17 A0A034WBF1 B4LR28 A0A0C9PU33 A0A1B0G0T1 A0A1A9Y1Y4 W8BXC3 A0A0A1WQJ9 A0A0K8SJY2 A0A0K8SII4 A0A1A9UIP7 A0A0Q9X7I6 A0A1A9Z2I7 B4PW74 B3NUY2 Q8IR45 B4R4W3
Ontologies
PANTHER
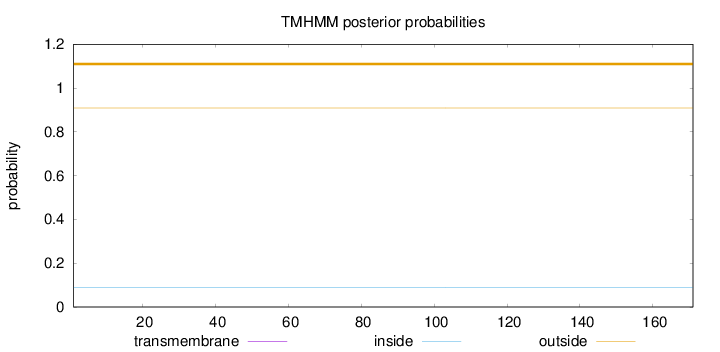
Topology
Subcellular location
Lysosome membrane
Length:
171
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09073
outside
1 - 171
Population Genetic Test Statistics
Pi
212.66204
Theta
188.165873
Tajima's D
0.404051
CLR
0
CSRT
0.48187590620469
Interpretation
Uncertain
