Gene
KWMTBOMO02453
Pre Gene Modal
BGIBMGA002730
Annotation
PREDICTED:_epidermal_retinal_dehydrogenase_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.051
Sequence
CDS
ATGTTACTACGACTCTATTCATTTTGCATTTTGATGGTGGATGTTGCTTGGGTTGTGTTCAATGCAATATGCGCAACATTTCAAGCTGCATATGAATTGTTCAGACCGCCACCATTGAAAACTGTCAGATTGGAAACTGCTATGGTCATTGGCTCCGGAAGAGGTGTTGGACGACAAATAGCTATGCAACTTGCAGATCTAGGGGCAATAGTACTGTGTATTGACATCAATCATCAGAATAATGAAGATACTGTGGACCACATCAAACTTAGAGGTGGATCAGCAGCCAGTTACATTTGTGATGTTACAAGAAGAGAGAAAATAGAAGAATTAGCAGCACAAATTAAGAAAGATGTTGGTTTTGTCAGCATGTTGTTCTATTGCTGTGGTATACCCAGCCCTAGGTCTTTAATGACACAACCACCTCAAGATATACACAAAACATTGGATTTGACATTGACATCATATTTCTGGCTCATAGACAACTTCTTACCAGAAATGAAAACCAGAGACCACGGGCACATTGTGGCTTTGACTTCTGTAGCTGGTCTCAGCTACATAAAAGATAAGATGCCGCTCAGTGTTGCTCAGTTTGCAGTGCAGGGGTTAGCGGAGTCGCTGATGGAGGATCTCAGAATAAATAAGATTGATAATGTGCATGTGACATTGATTCACATATATCCTTTCATCGTGAATGATGAGTATCACGATGTGAAATTAAGGATACCCAGCTATTTCGGGACTATATCACCTGTAGAAGCAGCATCATCGATCCTGGACAGCGTCAGGAGGAACTACCCGGAAGCATCAGTTCCAAAACATTTGTTATGTCTTGGTCATTTGTTACGTATTCTACCGCGCCGAGCAACAGTACTAATAAGGGACTTACTCGACACAGGTGTTGATTTTGCTTGA
Protein
MLLRLYSFCILMVDVAWVVFNAICATFQAAYELFRPPPLKTVRLETAMVIGSGRGVGRQIAMQLADLGAIVLCIDINHQNNEDTVDHIKLRGGSAASYICDVTRREKIEELAAQIKKDVGFVSMLFYCCGIPSPRSLMTQPPQDIHKTLDLTLTSYFWLIDNFLPEMKTRDHGHIVALTSVAGLSYIKDKMPLSVAQFAVQGLAESLMEDLRINKIDNVHVTLIHIYPFIVNDEYHDVKLRIPSYFGTISPVEAASSILDSVRRNYPEASVPKHLLCLGHLLRILPRRATVLIRDLLDTGVDFA
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
Q1HQ03
H9IZP5
A0A2W1BNJ1
A0A2A4IUS6
A0A2H1WW30
A0A194Q412
+ More
A0A212F2P0 S4PU03 A0A0L7L1Z5 A0A1Q3G5G9 A0A1Q3G5G5 A0A336K307 A0A2J7QD11 A0A0L7QVI3 F4W5L9 A0A087ZW96 A0A154P4Y5 A0A195ES88 A0A067QRK2 A0A158NSD5 A0A151JQG8 B0XHQ8 A0A0J7KMJ8 A0A2A3EMZ6 A0A023EMZ5 A0A1Q3FCP9 A0A182G8F2 A0A151WLN3 Q176M2 K7IU33 A0A182IMS1 A0A2M4AY87 A0A2P8YVI5 W5J961 A0A182LVD2 A0A084VAM0 A0A2M4AY85 A0A310SCI8 A0A026X000 U5EWJ5 E2AVV7 A0A182SCV0 A0A3L8D4F6 A0A182P9A1 A0A182YHJ7 A0A1S4GF86 Q7QC74 A0A2C9GRF4 A0A182N526 A0A151I2E0 A0A1J1I3C7 A0A182QLF7 A0A182R3V2 A0A182XHZ4 A0A182VG36 A0A182LF18 A0A182UEN5 A0A182VWP4 A0A182FKW1 A0A2M4AYX8 A0A182K4D0 A0A146MDQ4 B4Q0N3 A0A0K8T673 A0A146KSE4 A0A0A9XL74 B4IKW1 B3MRJ7 B4L823 A0A0N7Z933 B3NY14 K7IU30 A0A212EHJ0 B0WZ23 A0A1J1J5V5 A0A1Q3FQL1 T1IBK9 A0A224XGH6 Q9W3K9 Q8SWZ9 Q7PRW2 A0A0V0G980 A0A2C9GPE1 A0A1W4VXC3 A0A0M5J2W9 B4NEJ5 W5JLV1 A0A1L8DKW9 A0A182TE30 S4PKB0 A0A182L878 A0A182YH69 A0A182PSU8 A0A182UQG1 A0A182XBY7 A0A084VNW2 A0A182MHI6 A0A182W5M3 A0A023F371 A0A182RHK9
A0A212F2P0 S4PU03 A0A0L7L1Z5 A0A1Q3G5G9 A0A1Q3G5G5 A0A336K307 A0A2J7QD11 A0A0L7QVI3 F4W5L9 A0A087ZW96 A0A154P4Y5 A0A195ES88 A0A067QRK2 A0A158NSD5 A0A151JQG8 B0XHQ8 A0A0J7KMJ8 A0A2A3EMZ6 A0A023EMZ5 A0A1Q3FCP9 A0A182G8F2 A0A151WLN3 Q176M2 K7IU33 A0A182IMS1 A0A2M4AY87 A0A2P8YVI5 W5J961 A0A182LVD2 A0A084VAM0 A0A2M4AY85 A0A310SCI8 A0A026X000 U5EWJ5 E2AVV7 A0A182SCV0 A0A3L8D4F6 A0A182P9A1 A0A182YHJ7 A0A1S4GF86 Q7QC74 A0A2C9GRF4 A0A182N526 A0A151I2E0 A0A1J1I3C7 A0A182QLF7 A0A182R3V2 A0A182XHZ4 A0A182VG36 A0A182LF18 A0A182UEN5 A0A182VWP4 A0A182FKW1 A0A2M4AYX8 A0A182K4D0 A0A146MDQ4 B4Q0N3 A0A0K8T673 A0A146KSE4 A0A0A9XL74 B4IKW1 B3MRJ7 B4L823 A0A0N7Z933 B3NY14 K7IU30 A0A212EHJ0 B0WZ23 A0A1J1J5V5 A0A1Q3FQL1 T1IBK9 A0A224XGH6 Q9W3K9 Q8SWZ9 Q7PRW2 A0A0V0G980 A0A2C9GPE1 A0A1W4VXC3 A0A0M5J2W9 B4NEJ5 W5JLV1 A0A1L8DKW9 A0A182TE30 S4PKB0 A0A182L878 A0A182YH69 A0A182PSU8 A0A182UQG1 A0A182XBY7 A0A084VNW2 A0A182MHI6 A0A182W5M3 A0A023F371 A0A182RHK9
Pubmed
19121390
28756777
26354079
22118469
23622113
26227816
+ More
21719571 24845553 21347285 24945155 26483478 17510324 20075255 29403074 20920257 23761445 24438588 24508170 20798317 30249741 25244985 12364791 20966253 26823975 17994087 17550304 25401762 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14747013 17210077 25474469
21719571 24845553 21347285 24945155 26483478 17510324 20075255 29403074 20920257 23761445 24438588 24508170 20798317 30249741 25244985 12364791 20966253 26823975 17994087 17550304 25401762 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14747013 17210077 25474469
EMBL
DQ443249
ABF51338.1
BABH01014571
KZ149951
PZC76632.1
NWSH01006033
+ More
PCG63727.1 ODYU01011494 SOQ57258.1 KQ459465 KPJ00282.1 AGBW02010715 OWR47992.1 GAIX01011138 JAA81422.1 JTDY01003501 KOB69455.1 GEYN01000195 JAV44794.1 GEYN01000198 JAV44791.1 UFQS01000011 UFQT01000011 SSW97153.1 SSX17539.1 NEVH01015832 PNF26469.1 KQ414727 KOC62511.1 GL887655 EGI70516.1 KQ434819 KZC06947.1 KQ981993 KYN31036.1 KK853021 KDR12385.1 ADTU01024835 KQ978642 KYN29484.1 DS233191 EDS28565.1 LBMM01005480 KMQ91479.1 KZ288215 PBC32519.1 JXUM01007904 GAPW01002856 KQ560249 JAC10742.1 KXJ83576.1 GFDL01009777 JAV25268.1 JXUM01007910 KXJ83577.1 KQ982956 KYQ48799.1 CH477385 EAT42125.1 EAT42126.1 AAZX01009366 AAZX01016310 GGFK01012419 MBW45740.1 PYGN01000337 PSN48256.1 ADMH02002026 ETN59918.1 AXCM01007361 ATLV01004131 KE524190 KFB35014.1 GGFK01012420 MBW45741.1 KQ769068 OAD52993.1 KK107061 EZA61336.1 GANO01002927 JAB56944.1 GL443213 EFN62379.1 QOIP01000014 RLU15116.1 AAAB01008859 EAA08199.4 APCN01000376 KQ976543 KYM80964.1 CVRI01000039 CRK94699.1 AXCN02000175 GGFK01012670 MBW45991.1 GDHC01001477 JAQ17152.1 CM000162 EDX02304.1 GBRD01004802 GBRD01004801 JAG61019.1 GDHC01020487 JAP98141.1 GBHO01023203 JAG20401.1 CH480859 EDW52732.1 CH902622 EDV34402.1 CH933814 EDW05598.2 GDKW01001667 JAI54928.1 CH954180 EDV47535.1 AAZX01011807 AGBW02014879 OWR40948.1 DS232203 EDS37337.1 CVRI01000073 CRL07775.1 GFDL01005134 JAV29911.1 ACPB03013753 GFTR01004841 JAW11585.1 AE014298 BT044131 AAF46316.1 ACH92196.1 AY094928 AAM11281.1 AAAB01008846 EAA06477.6 GECL01001744 JAP04380.1 APCN01001259 CP012528 ALC48624.1 CH964239 EDW82164.2 ADMH02001200 ETN63754.1 GFDF01007094 JAV06990.1 GAIX01004600 JAA87960.1 ATLV01014931 KE524996 KFB39656.1 AXCM01002712 GBBI01003106 JAC15606.1
PCG63727.1 ODYU01011494 SOQ57258.1 KQ459465 KPJ00282.1 AGBW02010715 OWR47992.1 GAIX01011138 JAA81422.1 JTDY01003501 KOB69455.1 GEYN01000195 JAV44794.1 GEYN01000198 JAV44791.1 UFQS01000011 UFQT01000011 SSW97153.1 SSX17539.1 NEVH01015832 PNF26469.1 KQ414727 KOC62511.1 GL887655 EGI70516.1 KQ434819 KZC06947.1 KQ981993 KYN31036.1 KK853021 KDR12385.1 ADTU01024835 KQ978642 KYN29484.1 DS233191 EDS28565.1 LBMM01005480 KMQ91479.1 KZ288215 PBC32519.1 JXUM01007904 GAPW01002856 KQ560249 JAC10742.1 KXJ83576.1 GFDL01009777 JAV25268.1 JXUM01007910 KXJ83577.1 KQ982956 KYQ48799.1 CH477385 EAT42125.1 EAT42126.1 AAZX01009366 AAZX01016310 GGFK01012419 MBW45740.1 PYGN01000337 PSN48256.1 ADMH02002026 ETN59918.1 AXCM01007361 ATLV01004131 KE524190 KFB35014.1 GGFK01012420 MBW45741.1 KQ769068 OAD52993.1 KK107061 EZA61336.1 GANO01002927 JAB56944.1 GL443213 EFN62379.1 QOIP01000014 RLU15116.1 AAAB01008859 EAA08199.4 APCN01000376 KQ976543 KYM80964.1 CVRI01000039 CRK94699.1 AXCN02000175 GGFK01012670 MBW45991.1 GDHC01001477 JAQ17152.1 CM000162 EDX02304.1 GBRD01004802 GBRD01004801 JAG61019.1 GDHC01020487 JAP98141.1 GBHO01023203 JAG20401.1 CH480859 EDW52732.1 CH902622 EDV34402.1 CH933814 EDW05598.2 GDKW01001667 JAI54928.1 CH954180 EDV47535.1 AAZX01011807 AGBW02014879 OWR40948.1 DS232203 EDS37337.1 CVRI01000073 CRL07775.1 GFDL01005134 JAV29911.1 ACPB03013753 GFTR01004841 JAW11585.1 AE014298 BT044131 AAF46316.1 ACH92196.1 AY094928 AAM11281.1 AAAB01008846 EAA06477.6 GECL01001744 JAP04380.1 APCN01001259 CP012528 ALC48624.1 CH964239 EDW82164.2 ADMH02001200 ETN63754.1 GFDF01007094 JAV06990.1 GAIX01004600 JAA87960.1 ATLV01014931 KE524996 KFB39656.1 AXCM01002712 GBBI01003106 JAC15606.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000235965
+ More
UP000053825 UP000007755 UP000005203 UP000076502 UP000078541 UP000027135 UP000005205 UP000078492 UP000002320 UP000036403 UP000242457 UP000069940 UP000249989 UP000075809 UP000008820 UP000002358 UP000075880 UP000245037 UP000000673 UP000075883 UP000030765 UP000053097 UP000000311 UP000075901 UP000279307 UP000075885 UP000076408 UP000007062 UP000075840 UP000075884 UP000078540 UP000183832 UP000075886 UP000075900 UP000076407 UP000075903 UP000075882 UP000075902 UP000075920 UP000069272 UP000075881 UP000002282 UP000001292 UP000007801 UP000009192 UP000008711 UP000015103 UP000000803 UP000192221 UP000092553 UP000007798
UP000053825 UP000007755 UP000005203 UP000076502 UP000078541 UP000027135 UP000005205 UP000078492 UP000002320 UP000036403 UP000242457 UP000069940 UP000249989 UP000075809 UP000008820 UP000002358 UP000075880 UP000245037 UP000000673 UP000075883 UP000030765 UP000053097 UP000000311 UP000075901 UP000279307 UP000075885 UP000076408 UP000007062 UP000075840 UP000075884 UP000078540 UP000183832 UP000075886 UP000075900 UP000076407 UP000075903 UP000075882 UP000075902 UP000075920 UP000069272 UP000075881 UP000002282 UP000001292 UP000007801 UP000009192 UP000008711 UP000015103 UP000000803 UP000192221 UP000092553 UP000007798
Interpro
Gene 3D
ProteinModelPortal
Q1HQ03
H9IZP5
A0A2W1BNJ1
A0A2A4IUS6
A0A2H1WW30
A0A194Q412
+ More
A0A212F2P0 S4PU03 A0A0L7L1Z5 A0A1Q3G5G9 A0A1Q3G5G5 A0A336K307 A0A2J7QD11 A0A0L7QVI3 F4W5L9 A0A087ZW96 A0A154P4Y5 A0A195ES88 A0A067QRK2 A0A158NSD5 A0A151JQG8 B0XHQ8 A0A0J7KMJ8 A0A2A3EMZ6 A0A023EMZ5 A0A1Q3FCP9 A0A182G8F2 A0A151WLN3 Q176M2 K7IU33 A0A182IMS1 A0A2M4AY87 A0A2P8YVI5 W5J961 A0A182LVD2 A0A084VAM0 A0A2M4AY85 A0A310SCI8 A0A026X000 U5EWJ5 E2AVV7 A0A182SCV0 A0A3L8D4F6 A0A182P9A1 A0A182YHJ7 A0A1S4GF86 Q7QC74 A0A2C9GRF4 A0A182N526 A0A151I2E0 A0A1J1I3C7 A0A182QLF7 A0A182R3V2 A0A182XHZ4 A0A182VG36 A0A182LF18 A0A182UEN5 A0A182VWP4 A0A182FKW1 A0A2M4AYX8 A0A182K4D0 A0A146MDQ4 B4Q0N3 A0A0K8T673 A0A146KSE4 A0A0A9XL74 B4IKW1 B3MRJ7 B4L823 A0A0N7Z933 B3NY14 K7IU30 A0A212EHJ0 B0WZ23 A0A1J1J5V5 A0A1Q3FQL1 T1IBK9 A0A224XGH6 Q9W3K9 Q8SWZ9 Q7PRW2 A0A0V0G980 A0A2C9GPE1 A0A1W4VXC3 A0A0M5J2W9 B4NEJ5 W5JLV1 A0A1L8DKW9 A0A182TE30 S4PKB0 A0A182L878 A0A182YH69 A0A182PSU8 A0A182UQG1 A0A182XBY7 A0A084VNW2 A0A182MHI6 A0A182W5M3 A0A023F371 A0A182RHK9
A0A212F2P0 S4PU03 A0A0L7L1Z5 A0A1Q3G5G9 A0A1Q3G5G5 A0A336K307 A0A2J7QD11 A0A0L7QVI3 F4W5L9 A0A087ZW96 A0A154P4Y5 A0A195ES88 A0A067QRK2 A0A158NSD5 A0A151JQG8 B0XHQ8 A0A0J7KMJ8 A0A2A3EMZ6 A0A023EMZ5 A0A1Q3FCP9 A0A182G8F2 A0A151WLN3 Q176M2 K7IU33 A0A182IMS1 A0A2M4AY87 A0A2P8YVI5 W5J961 A0A182LVD2 A0A084VAM0 A0A2M4AY85 A0A310SCI8 A0A026X000 U5EWJ5 E2AVV7 A0A182SCV0 A0A3L8D4F6 A0A182P9A1 A0A182YHJ7 A0A1S4GF86 Q7QC74 A0A2C9GRF4 A0A182N526 A0A151I2E0 A0A1J1I3C7 A0A182QLF7 A0A182R3V2 A0A182XHZ4 A0A182VG36 A0A182LF18 A0A182UEN5 A0A182VWP4 A0A182FKW1 A0A2M4AYX8 A0A182K4D0 A0A146MDQ4 B4Q0N3 A0A0K8T673 A0A146KSE4 A0A0A9XL74 B4IKW1 B3MRJ7 B4L823 A0A0N7Z933 B3NY14 K7IU30 A0A212EHJ0 B0WZ23 A0A1J1J5V5 A0A1Q3FQL1 T1IBK9 A0A224XGH6 Q9W3K9 Q8SWZ9 Q7PRW2 A0A0V0G980 A0A2C9GPE1 A0A1W4VXC3 A0A0M5J2W9 B4NEJ5 W5JLV1 A0A1L8DKW9 A0A182TE30 S4PKB0 A0A182L878 A0A182YH69 A0A182PSU8 A0A182UQG1 A0A182XBY7 A0A084VNW2 A0A182MHI6 A0A182W5M3 A0A023F371 A0A182RHK9
PDB
1YB1
E-value=4.07133e-22,
Score=257
Ontologies
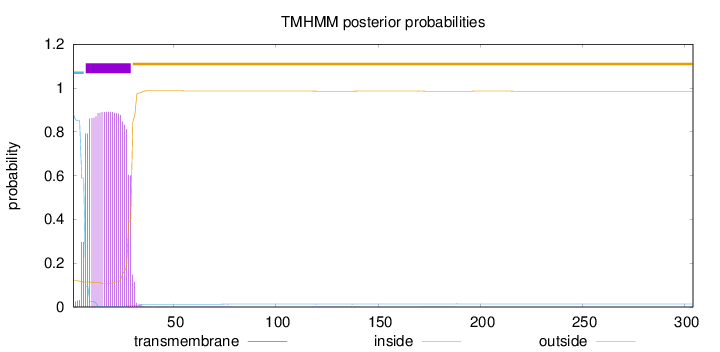
Topology
Length:
304
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.4272099999999
Exp number, first 60 AAs:
20.3826
Total prob of N-in:
0.88010
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 304
Population Genetic Test Statistics
Pi
211.431169
Theta
193.011212
Tajima's D
0.238224
CLR
0.340932
CSRT
0.437578121093945
Interpretation
Uncertain
