Gene
KWMTBOMO02450 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002559
Annotation
ubiquitin_conjugating_enzyme_E2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.354 Mitochondrial Reliability : 1.649
Sequence
CDS
ATGGCAGCCCTACCACGTAGAATAATCAAAGAGACACAGCGATTGATGCAGGAGCCGGTGCCCGGGATCAGCGCAGTACCTAGCGAGACAAACGCCCGTTACTTCCACGTTATCGTGACGGGCCCAGAGGACTCGCCTTTCGAAGGAGGACTGTTCAAATTAGAATTATTTCTACCGGAGGATTACCCCATGTCGGCACCTAAGGTTAGGTTTATCACGAAAATATATCATCCTAATATAGACCGACTGGGCCGCATTTGTTTGGACATTCTTAAAGATAAATGGAGTCCAGCTCTACAAATACGCACGGTGCTACTGTCTATTCAGGCTCTGCTGTCAGCACCTAACCCTGACGATCCGCTGGCGAACGACGTTGCCGAATTGTGGAAGGTGAACGAGAGTGAAGCTATTAGGAACGCGAAGGAGTGGACGCGGAGATACGCCATGGACAACTGA
Protein
MAALPRRIIKETQRLMQEPVPGISAVPSETNARYFHVIVTGPEDSPFEGGLFKLELFLPEDYPMSAPKVRFITKIYHPNIDRLGRICLDILKDKWSPALQIRTVLLSIQALLSAPNPDDPLANDVAELWKVNESEAIRNAKEWTRRYAMDN
Summary
Similarity
Belongs to the ubiquitin-conjugating enzyme family.
Uniprot
A0A2W1BVF9
A0A2A4J7D2
A0A0N0PBU7
A0A1E1WS29
A0A2H1W6K4
Q1HQ36
+ More
S4PGI9 A0A212FG39 A0A0J7K0U7 A0A151IEG3 E9IKM7 A0A0L7R2F7 A0A154PKP5 A0A151HYB9 A0A2A3E6A5 V9ICY9 A0A0M8ZQK4 A0A088A3B1 E2B9H8 A0A158NSE8 E2AX53 A0A151WXB7 A0A026W879 D6W8B6 A0A1B6G1Y6 A0A1B6HU24 A0A1B6L583 A0A195ES24 A0A0C9PY78 A0A0L7LR21 A0A0C9RHQ9 A0A310SQT3 A0A2J7R3W4 E0W2J0 A0A1S4ES97 V5G6R4 N6U6Q5 A0A067QUW4 A0A2Z5TTP2 A0A2P8YHJ6 A0A232FDW7 F4W5N4 K7J1X2 A0A1I9WLE8 A0A1J1IGY6 A0A1W4WNT0 A0A0K8TP38 A0A1Y1LVM8 T1RTE4 A0A1B0DKC3 A0A1B6EGV3 A0A1J1IM03 A0A182HNZ7 A0A2R7WF44 A0A0A9YCI0 A0A1L8DT45 U5EZB5 T1GMN4 T1DIY7 Q16FT8 Q5MIQ3 A0A0P4VUM3 T1I671 A0A224XYW9 A0A0V0G7B3 A0A023F5U5 A0A069DPG9 A0A1Q3FH14 B0WH99 A0A084WKU4 A0A182IYN3 A0A182XEX1 A0A182VEY4 A0A182YLB5 A0A182MJR9 A0A182P8Q7 A0A182R022 A0A182N1R1 A0A182JZX2 A0A182WAH8 A0A182LBM1 Q7Q9K0 A0A182RX13 A0A182F9N6 T1DNP1 A0A2M3ZHB2 A0A2M4AIQ1 A0A2M4C2N2 W5JQ37 B3MZT3 A0A1D2N6A8 A0A1D2NH33 A0A336LSQ3 A0A0M4F9M7 A0A2S2NWN4 A0A1W4VUA9 A0A3B0JXN2 B4H0K9 B4Q2P5 B4IG91
S4PGI9 A0A212FG39 A0A0J7K0U7 A0A151IEG3 E9IKM7 A0A0L7R2F7 A0A154PKP5 A0A151HYB9 A0A2A3E6A5 V9ICY9 A0A0M8ZQK4 A0A088A3B1 E2B9H8 A0A158NSE8 E2AX53 A0A151WXB7 A0A026W879 D6W8B6 A0A1B6G1Y6 A0A1B6HU24 A0A1B6L583 A0A195ES24 A0A0C9PY78 A0A0L7LR21 A0A0C9RHQ9 A0A310SQT3 A0A2J7R3W4 E0W2J0 A0A1S4ES97 V5G6R4 N6U6Q5 A0A067QUW4 A0A2Z5TTP2 A0A2P8YHJ6 A0A232FDW7 F4W5N4 K7J1X2 A0A1I9WLE8 A0A1J1IGY6 A0A1W4WNT0 A0A0K8TP38 A0A1Y1LVM8 T1RTE4 A0A1B0DKC3 A0A1B6EGV3 A0A1J1IM03 A0A182HNZ7 A0A2R7WF44 A0A0A9YCI0 A0A1L8DT45 U5EZB5 T1GMN4 T1DIY7 Q16FT8 Q5MIQ3 A0A0P4VUM3 T1I671 A0A224XYW9 A0A0V0G7B3 A0A023F5U5 A0A069DPG9 A0A1Q3FH14 B0WH99 A0A084WKU4 A0A182IYN3 A0A182XEX1 A0A182VEY4 A0A182YLB5 A0A182MJR9 A0A182P8Q7 A0A182R022 A0A182N1R1 A0A182JZX2 A0A182WAH8 A0A182LBM1 Q7Q9K0 A0A182RX13 A0A182F9N6 T1DNP1 A0A2M3ZHB2 A0A2M4AIQ1 A0A2M4C2N2 W5JQ37 B3MZT3 A0A1D2N6A8 A0A1D2NH33 A0A336LSQ3 A0A0M4F9M7 A0A2S2NWN4 A0A1W4VUA9 A0A3B0JXN2 B4H0K9 B4Q2P5 B4IG91
Pubmed
28756777
26354079
19121390
23622113
22118469
21282665
+ More
20798317 21347285 24508170 30249741 18362917 19820115 26227816 20566863 23537049 24845553 26760975 29403074 28648823 21719571 20075255 27538518 26369729 28004739 25401762 26823975 24330624 17510324 17244540 24945155 26483478 27129103 25474469 26334808 24438588 25244985 20966253 12364791 14747013 17210077 20920257 23761445 17994087 18057021 27289101 17550304
20798317 21347285 24508170 30249741 18362917 19820115 26227816 20566863 23537049 24845553 26760975 29403074 28648823 21719571 20075255 27538518 26369729 28004739 25401762 26823975 24330624 17510324 17244540 24945155 26483478 27129103 25474469 26334808 24438588 25244985 20966253 12364791 14747013 17210077 20920257 23761445 17994087 18057021 27289101 17550304
EMBL
KZ149951
PZC76636.1
NWSH01002604
PCG67871.1
KQ460796
KPJ12159.1
+ More
GDQN01001234 JAT89820.1 ODYU01006524 SOQ48452.1 BABH01014585 DQ443216 ABF51305.1 GAIX01003642 JAA88918.1 AGBW02008722 OWR52698.1 LBMM01018398 KMQ83796.1 KQ977894 KYM98957.1 GL764021 EFZ18873.1 KQ414666 KOC65013.1 KQ434938 KZC12044.1 KQ976730 KYM76470.1 KZ288354 PBC27238.1 JR039066 AEY58517.1 KQ435900 KOX69095.1 GL446527 EFN87661.1 ADTU01024876 ADTU01024877 GL443520 EFN62003.1 KQ982669 KYQ52456.1 KK107347 QOIP01000007 EZA52265.1 RLU20330.1 KQ971307 EFA10928.1 GECZ01013312 JAS56457.1 GECU01029530 JAS78176.1 GEBQ01021147 GEBQ01000152 JAT18830.1 JAT39825.1 KQ981993 KYN31055.1 GBYB01006493 JAG76260.1 JTDY01000276 KOB77948.1 GBYB01006491 GBYB01006492 JAG76258.1 JAG76259.1 KQ760133 OAD61782.1 NEVH01007818 PNF35524.1 DS235878 EEB19846.1 GALX01002691 JAB65775.1 APGK01047231 KB741077 KB631792 ENN74247.1 ERL86125.1 KK852921 KDR13781.1 FX985838 BBA93725.1 PYGN01000591 PSN43712.1 NNAY01000355 OXU28951.1 GL887655 EGI70531.1 KU932332 APA33968.1 CVRI01000048 CRK99034.1 GDAI01001494 JAI16109.1 GEZM01045881 JAV77642.1 KC569748 AGO46417.1 AJVK01066813 GEDC01000194 JAS37104.1 CVRI01000051 CRK99497.1 APCN01002820 KK854679 PTY17851.1 GBHO01014791 GBRD01013307 GDHC01007926 JAG28813.1 JAG52519.1 JAQ10703.1 GFDF01004458 JAV09626.1 GANO01001092 JAB58779.1 CAQQ02026378 GALA01000656 JAA94196.1 CH478382 EAT33096.1 EAT33097.1 JXUM01125342 JXUM01145687 AY826157 GAPW01005469 KQ570031 KQ566934 AAV90729.1 JAC08129.1 KXJ68469.1 KXJ69718.1 GDKW01000866 JAI55729.1 ACPB03023977 GFTR01002616 JAW13810.1 GECL01002802 JAP03322.1 GBBI01002157 JAC16555.1 GBGD01003308 JAC85581.1 GFDL01008220 JAV26825.1 DS231933 EDS27557.1 ATLV01024147 KE525350 KFB50838.1 AXCP01006002 AXCM01001476 AXCN02000987 AAAB01008900 EAA09423.2 GAMD01002680 JAA98910.1 GGFM01007173 MBW27924.1 GGFK01007339 MBW40660.1 GGFJ01010280 MBW59421.1 ADMH02000800 GGFL01004234 ETN65000.1 MBW68412.1 CH902635 EDV33884.1 KPU74824.1 KPU74825.1 KPU74826.1 LJIJ01000186 ODN00787.1 LJIJ01000044 ODN04405.1 UFQS01000096 UFQT01000096 SSW99363.1 SSX19743.1 CP012528 ALC48957.1 GGMR01008843 MBY21462.1 OUUW01000011 SPP86814.1 CH479200 EDW29804.1 CM000162 EDX01639.1 CH480835 EDW48825.1
GDQN01001234 JAT89820.1 ODYU01006524 SOQ48452.1 BABH01014585 DQ443216 ABF51305.1 GAIX01003642 JAA88918.1 AGBW02008722 OWR52698.1 LBMM01018398 KMQ83796.1 KQ977894 KYM98957.1 GL764021 EFZ18873.1 KQ414666 KOC65013.1 KQ434938 KZC12044.1 KQ976730 KYM76470.1 KZ288354 PBC27238.1 JR039066 AEY58517.1 KQ435900 KOX69095.1 GL446527 EFN87661.1 ADTU01024876 ADTU01024877 GL443520 EFN62003.1 KQ982669 KYQ52456.1 KK107347 QOIP01000007 EZA52265.1 RLU20330.1 KQ971307 EFA10928.1 GECZ01013312 JAS56457.1 GECU01029530 JAS78176.1 GEBQ01021147 GEBQ01000152 JAT18830.1 JAT39825.1 KQ981993 KYN31055.1 GBYB01006493 JAG76260.1 JTDY01000276 KOB77948.1 GBYB01006491 GBYB01006492 JAG76258.1 JAG76259.1 KQ760133 OAD61782.1 NEVH01007818 PNF35524.1 DS235878 EEB19846.1 GALX01002691 JAB65775.1 APGK01047231 KB741077 KB631792 ENN74247.1 ERL86125.1 KK852921 KDR13781.1 FX985838 BBA93725.1 PYGN01000591 PSN43712.1 NNAY01000355 OXU28951.1 GL887655 EGI70531.1 KU932332 APA33968.1 CVRI01000048 CRK99034.1 GDAI01001494 JAI16109.1 GEZM01045881 JAV77642.1 KC569748 AGO46417.1 AJVK01066813 GEDC01000194 JAS37104.1 CVRI01000051 CRK99497.1 APCN01002820 KK854679 PTY17851.1 GBHO01014791 GBRD01013307 GDHC01007926 JAG28813.1 JAG52519.1 JAQ10703.1 GFDF01004458 JAV09626.1 GANO01001092 JAB58779.1 CAQQ02026378 GALA01000656 JAA94196.1 CH478382 EAT33096.1 EAT33097.1 JXUM01125342 JXUM01145687 AY826157 GAPW01005469 KQ570031 KQ566934 AAV90729.1 JAC08129.1 KXJ68469.1 KXJ69718.1 GDKW01000866 JAI55729.1 ACPB03023977 GFTR01002616 JAW13810.1 GECL01002802 JAP03322.1 GBBI01002157 JAC16555.1 GBGD01003308 JAC85581.1 GFDL01008220 JAV26825.1 DS231933 EDS27557.1 ATLV01024147 KE525350 KFB50838.1 AXCP01006002 AXCM01001476 AXCN02000987 AAAB01008900 EAA09423.2 GAMD01002680 JAA98910.1 GGFM01007173 MBW27924.1 GGFK01007339 MBW40660.1 GGFJ01010280 MBW59421.1 ADMH02000800 GGFL01004234 ETN65000.1 MBW68412.1 CH902635 EDV33884.1 KPU74824.1 KPU74825.1 KPU74826.1 LJIJ01000186 ODN00787.1 LJIJ01000044 ODN04405.1 UFQS01000096 UFQT01000096 SSW99363.1 SSX19743.1 CP012528 ALC48957.1 GGMR01008843 MBY21462.1 OUUW01000011 SPP86814.1 CH479200 EDW29804.1 CM000162 EDX01639.1 CH480835 EDW48825.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000007151
UP000036403
UP000078542
+ More
UP000053825 UP000076502 UP000078540 UP000242457 UP000053105 UP000005203 UP000008237 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000007266 UP000078541 UP000037510 UP000235965 UP000009046 UP000079169 UP000019118 UP000030742 UP000027135 UP000245037 UP000215335 UP000007755 UP000002358 UP000183832 UP000192223 UP000092462 UP000075840 UP000015102 UP000008820 UP000069940 UP000249989 UP000015103 UP000002320 UP000030765 UP000075880 UP000076407 UP000075903 UP000076408 UP000075883 UP000075885 UP000075886 UP000075884 UP000075881 UP000075920 UP000075882 UP000007062 UP000075900 UP000069272 UP000000673 UP000007801 UP000094527 UP000092553 UP000192221 UP000268350 UP000008744 UP000002282 UP000001292
UP000053825 UP000076502 UP000078540 UP000242457 UP000053105 UP000005203 UP000008237 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000007266 UP000078541 UP000037510 UP000235965 UP000009046 UP000079169 UP000019118 UP000030742 UP000027135 UP000245037 UP000215335 UP000007755 UP000002358 UP000183832 UP000192223 UP000092462 UP000075840 UP000015102 UP000008820 UP000069940 UP000249989 UP000015103 UP000002320 UP000030765 UP000075880 UP000076407 UP000075903 UP000076408 UP000075883 UP000075885 UP000075886 UP000075884 UP000075881 UP000075920 UP000075882 UP000007062 UP000075900 UP000069272 UP000000673 UP000007801 UP000094527 UP000092553 UP000192221 UP000268350 UP000008744 UP000002282 UP000001292
PRIDE
Pfam
PF00179 UQ_con
Interpro
SUPFAM
SSF54495
SSF54495
Gene 3D
CDD
ProteinModelPortal
A0A2W1BVF9
A0A2A4J7D2
A0A0N0PBU7
A0A1E1WS29
A0A2H1W6K4
Q1HQ36
+ More
S4PGI9 A0A212FG39 A0A0J7K0U7 A0A151IEG3 E9IKM7 A0A0L7R2F7 A0A154PKP5 A0A151HYB9 A0A2A3E6A5 V9ICY9 A0A0M8ZQK4 A0A088A3B1 E2B9H8 A0A158NSE8 E2AX53 A0A151WXB7 A0A026W879 D6W8B6 A0A1B6G1Y6 A0A1B6HU24 A0A1B6L583 A0A195ES24 A0A0C9PY78 A0A0L7LR21 A0A0C9RHQ9 A0A310SQT3 A0A2J7R3W4 E0W2J0 A0A1S4ES97 V5G6R4 N6U6Q5 A0A067QUW4 A0A2Z5TTP2 A0A2P8YHJ6 A0A232FDW7 F4W5N4 K7J1X2 A0A1I9WLE8 A0A1J1IGY6 A0A1W4WNT0 A0A0K8TP38 A0A1Y1LVM8 T1RTE4 A0A1B0DKC3 A0A1B6EGV3 A0A1J1IM03 A0A182HNZ7 A0A2R7WF44 A0A0A9YCI0 A0A1L8DT45 U5EZB5 T1GMN4 T1DIY7 Q16FT8 Q5MIQ3 A0A0P4VUM3 T1I671 A0A224XYW9 A0A0V0G7B3 A0A023F5U5 A0A069DPG9 A0A1Q3FH14 B0WH99 A0A084WKU4 A0A182IYN3 A0A182XEX1 A0A182VEY4 A0A182YLB5 A0A182MJR9 A0A182P8Q7 A0A182R022 A0A182N1R1 A0A182JZX2 A0A182WAH8 A0A182LBM1 Q7Q9K0 A0A182RX13 A0A182F9N6 T1DNP1 A0A2M3ZHB2 A0A2M4AIQ1 A0A2M4C2N2 W5JQ37 B3MZT3 A0A1D2N6A8 A0A1D2NH33 A0A336LSQ3 A0A0M4F9M7 A0A2S2NWN4 A0A1W4VUA9 A0A3B0JXN2 B4H0K9 B4Q2P5 B4IG91
S4PGI9 A0A212FG39 A0A0J7K0U7 A0A151IEG3 E9IKM7 A0A0L7R2F7 A0A154PKP5 A0A151HYB9 A0A2A3E6A5 V9ICY9 A0A0M8ZQK4 A0A088A3B1 E2B9H8 A0A158NSE8 E2AX53 A0A151WXB7 A0A026W879 D6W8B6 A0A1B6G1Y6 A0A1B6HU24 A0A1B6L583 A0A195ES24 A0A0C9PY78 A0A0L7LR21 A0A0C9RHQ9 A0A310SQT3 A0A2J7R3W4 E0W2J0 A0A1S4ES97 V5G6R4 N6U6Q5 A0A067QUW4 A0A2Z5TTP2 A0A2P8YHJ6 A0A232FDW7 F4W5N4 K7J1X2 A0A1I9WLE8 A0A1J1IGY6 A0A1W4WNT0 A0A0K8TP38 A0A1Y1LVM8 T1RTE4 A0A1B0DKC3 A0A1B6EGV3 A0A1J1IM03 A0A182HNZ7 A0A2R7WF44 A0A0A9YCI0 A0A1L8DT45 U5EZB5 T1GMN4 T1DIY7 Q16FT8 Q5MIQ3 A0A0P4VUM3 T1I671 A0A224XYW9 A0A0V0G7B3 A0A023F5U5 A0A069DPG9 A0A1Q3FH14 B0WH99 A0A084WKU4 A0A182IYN3 A0A182XEX1 A0A182VEY4 A0A182YLB5 A0A182MJR9 A0A182P8Q7 A0A182R022 A0A182N1R1 A0A182JZX2 A0A182WAH8 A0A182LBM1 Q7Q9K0 A0A182RX13 A0A182F9N6 T1DNP1 A0A2M3ZHB2 A0A2M4AIQ1 A0A2M4C2N2 W5JQ37 B3MZT3 A0A1D2N6A8 A0A1D2NH33 A0A336LSQ3 A0A0M4F9M7 A0A2S2NWN4 A0A1W4VUA9 A0A3B0JXN2 B4H0K9 B4Q2P5 B4IG91
PDB
4IP3
E-value=2.68322e-58,
Score=565
Ontologies
PATHWAY
GO
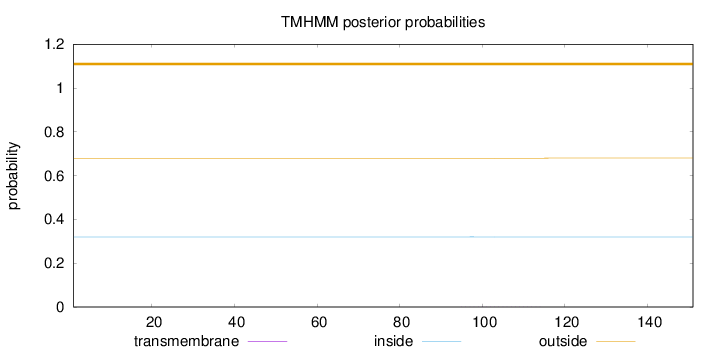
Topology
Length:
151
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00527
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.32025
outside
1 - 151
Population Genetic Test Statistics
Pi
168.26074
Theta
190.285796
Tajima's D
-0.563265
CLR
7.054356
CSRT
0.230888455577221
Interpretation
Uncertain
