Gene
KWMTBOMO02445 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013504
Annotation
hypothetical_protein_KGM_09857_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 2.191
Sequence
CDS
ATGGGGAGTTCGCAAAGTGCGCGGAAGCTTAGCGTGGATAATGAAAATGCTATTCAAGTGTCTCAAGAAGCCATAGACCGAATACAGGCTCAACTAAACGCGAAACAAGCGGAACAACCAAAACCAGTTCCACAGTATCTTCCTCCACCTCCATACTATGATCAGCGAAAACAACAGGAGCATGCTGCTGAAGAAGCATACTGGACTCGACGCATTGAAAACTTAAAAAGAGTACATGAAAAAATCAATAGTGGCATGGAATTAGAATATCAGAAAACACTGGAAGAGACAAATGAGCTATTTAATTTCATAAATAATGATAAAAATGTGAATAAACTGCCTCCATGCCAAGAAGAAAAAGAAAAGGTACTTCAGTGTTACAGTACAAATGCGGACAAGTCTCTCCTTTGTTCAGTGTTAGTGAATCAGTTCAATGAGTGCGTATGCCGAAGCAGGGTGGCCGCTATCACAGCTTCTTGA
Protein
MGSSQSARKLSVDNENAIQVSQEAIDRIQAQLNAKQAEQPKPVPQYLPPPPYYDQRKQQEHAAEEAYWTRRIENLKRVHEKINSGMELEYQKTLEETNELFNFINNDKNVNKLPPCQEEKEKVLQCYSTNADKSLLCSVLVNQFNECVCRSRVAAITAS
Summary
Uniprot
H9JVE1
A0A2H1W8Q0
S4PA02
A0A212FI48
I4DNL1
A0A2A4JXP0
+ More
A0A2W1BH56 A0A0L7LT81 A0A1B6DH61 A0A1W4WML9 D6WIF7 A0A154PBQ8 R4WSF3 A0A310SG58 A0A2A3EMB2 V9ILP4 A0A088AVC8 A0A026W0D7 V5GRH9 Q17DU1 B0WQV9 A0A1S4F6K4 A0A1Q3FG85 N6TSI5 A0A1S3DN68 A0A1B6H729 A0A067QNV7 A0A1J1ITE6 A0A1B6GRK2 A0A1Y1L4F2 A0A3Q0J661 A0A2R7W9U0 A0A2H1VNF7 F4WLT5 A0A1B6LRD8 A0A2J7RNC6 U5EQ92 A0A069DQ23 A0A0P4W4H2 R4G424 A0A158NWV6 A0A224XZ29 A0A151IT78 E2BQM6 A0A0V0G5Z6 A0A195FX06 R4UWX6
A0A2W1BH56 A0A0L7LT81 A0A1B6DH61 A0A1W4WML9 D6WIF7 A0A154PBQ8 R4WSF3 A0A310SG58 A0A2A3EMB2 V9ILP4 A0A088AVC8 A0A026W0D7 V5GRH9 Q17DU1 B0WQV9 A0A1S4F6K4 A0A1Q3FG85 N6TSI5 A0A1S3DN68 A0A1B6H729 A0A067QNV7 A0A1J1ITE6 A0A1B6GRK2 A0A1Y1L4F2 A0A3Q0J661 A0A2R7W9U0 A0A2H1VNF7 F4WLT5 A0A1B6LRD8 A0A2J7RNC6 U5EQ92 A0A069DQ23 A0A0P4W4H2 R4G424 A0A158NWV6 A0A224XZ29 A0A151IT78 E2BQM6 A0A0V0G5Z6 A0A195FX06 R4UWX6
Pubmed
EMBL
BABH01036027
ODYU01007043
SOQ49448.1
GAIX01003474
JAA89086.1
AGBW02008434
+ More
OWR53408.1 AK402990 KQ459465 BAM19501.1 KPJ00290.1 NWSH01000385 PCG76781.1 KZ150213 PZC72146.1 JTDY01000137 KOB78663.1 GEDC01012272 GEDC01005080 JAS25026.1 JAS32218.1 KQ971334 EEZ99676.1 KQ434869 KZC09339.1 AK417632 BAN20847.1 KQ761546 OAD57411.1 KZ288210 PBC32943.1 JR053704 AEY61995.1 KK107510 EZA49507.1 GALX01005598 JAB62868.1 CH477291 EAT44571.1 DS232046 EDS33039.1 GFDL01008490 JAV26555.1 APGK01053605 KB741231 KB631770 ENN72220.1 ERL86006.1 GECU01037285 JAS70421.1 KK853110 KDR11231.1 CVRI01000059 CRL03479.1 GECZ01004708 JAS65061.1 GEZM01066861 JAV67678.1 KK854356 PTY15035.1 ODYU01003478 SOQ42327.1 GL888216 EGI64817.1 GEBQ01013724 JAT26253.1 NEVH01002545 PNF42322.1 GANO01004398 JAB55473.1 GBGD01003108 JAC85781.1 GDKW01000362 JAI56233.1 ACPB03021527 GAHY01001715 JAA75795.1 ADTU01028644 GFTR01003125 JAW13301.1 KQ981033 KYN10173.1 GL449784 EFN81991.1 GECL01002641 JAP03483.1 KQ981204 KYN44961.1 KC740987 AGM32811.1
OWR53408.1 AK402990 KQ459465 BAM19501.1 KPJ00290.1 NWSH01000385 PCG76781.1 KZ150213 PZC72146.1 JTDY01000137 KOB78663.1 GEDC01012272 GEDC01005080 JAS25026.1 JAS32218.1 KQ971334 EEZ99676.1 KQ434869 KZC09339.1 AK417632 BAN20847.1 KQ761546 OAD57411.1 KZ288210 PBC32943.1 JR053704 AEY61995.1 KK107510 EZA49507.1 GALX01005598 JAB62868.1 CH477291 EAT44571.1 DS232046 EDS33039.1 GFDL01008490 JAV26555.1 APGK01053605 KB741231 KB631770 ENN72220.1 ERL86006.1 GECU01037285 JAS70421.1 KK853110 KDR11231.1 CVRI01000059 CRL03479.1 GECZ01004708 JAS65061.1 GEZM01066861 JAV67678.1 KK854356 PTY15035.1 ODYU01003478 SOQ42327.1 GL888216 EGI64817.1 GEBQ01013724 JAT26253.1 NEVH01002545 PNF42322.1 GANO01004398 JAB55473.1 GBGD01003108 JAC85781.1 GDKW01000362 JAI56233.1 ACPB03021527 GAHY01001715 JAA75795.1 ADTU01028644 GFTR01003125 JAW13301.1 KQ981033 KYN10173.1 GL449784 EFN81991.1 GECL01002641 JAP03483.1 KQ981204 KYN44961.1 KC740987 AGM32811.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000037510
UP000192223
+ More
UP000007266 UP000076502 UP000242457 UP000005203 UP000053097 UP000008820 UP000002320 UP000019118 UP000030742 UP000079169 UP000027135 UP000183832 UP000007755 UP000235965 UP000015103 UP000005205 UP000078492 UP000008237 UP000078541
UP000007266 UP000076502 UP000242457 UP000005203 UP000053097 UP000008820 UP000002320 UP000019118 UP000030742 UP000079169 UP000027135 UP000183832 UP000007755 UP000235965 UP000015103 UP000005205 UP000078492 UP000008237 UP000078541
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JVE1
A0A2H1W8Q0
S4PA02
A0A212FI48
I4DNL1
A0A2A4JXP0
+ More
A0A2W1BH56 A0A0L7LT81 A0A1B6DH61 A0A1W4WML9 D6WIF7 A0A154PBQ8 R4WSF3 A0A310SG58 A0A2A3EMB2 V9ILP4 A0A088AVC8 A0A026W0D7 V5GRH9 Q17DU1 B0WQV9 A0A1S4F6K4 A0A1Q3FG85 N6TSI5 A0A1S3DN68 A0A1B6H729 A0A067QNV7 A0A1J1ITE6 A0A1B6GRK2 A0A1Y1L4F2 A0A3Q0J661 A0A2R7W9U0 A0A2H1VNF7 F4WLT5 A0A1B6LRD8 A0A2J7RNC6 U5EQ92 A0A069DQ23 A0A0P4W4H2 R4G424 A0A158NWV6 A0A224XZ29 A0A151IT78 E2BQM6 A0A0V0G5Z6 A0A195FX06 R4UWX6
A0A2W1BH56 A0A0L7LT81 A0A1B6DH61 A0A1W4WML9 D6WIF7 A0A154PBQ8 R4WSF3 A0A310SG58 A0A2A3EMB2 V9ILP4 A0A088AVC8 A0A026W0D7 V5GRH9 Q17DU1 B0WQV9 A0A1S4F6K4 A0A1Q3FG85 N6TSI5 A0A1S3DN68 A0A1B6H729 A0A067QNV7 A0A1J1ITE6 A0A1B6GRK2 A0A1Y1L4F2 A0A3Q0J661 A0A2R7W9U0 A0A2H1VNF7 F4WLT5 A0A1B6LRD8 A0A2J7RNC6 U5EQ92 A0A069DQ23 A0A0P4W4H2 R4G424 A0A158NWV6 A0A224XZ29 A0A151IT78 E2BQM6 A0A0V0G5Z6 A0A195FX06 R4UWX6
Ontologies
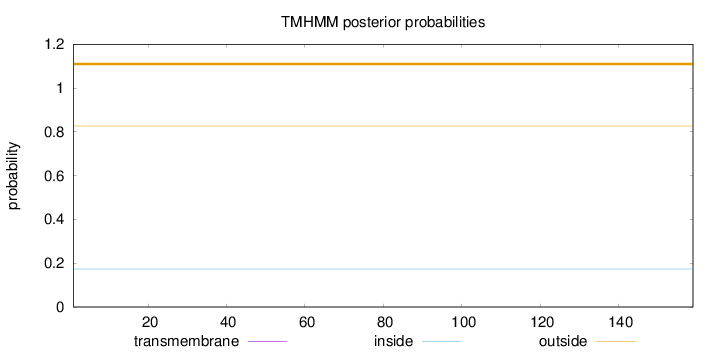
Topology
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00023
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17344
outside
1 - 159
Population Genetic Test Statistics
Pi
237.424573
Theta
201.266822
Tajima's D
0.65164
CLR
0.65975
CSRT
0.565521723913804
Interpretation
Uncertain
