Gene
KWMTBOMO02441
Pre Gene Modal
BGIBMGA013502
Annotation
PREDICTED:_protein_pigeon_[Bombyx_mori]
Full name
Protein pigeon
Alternative Name
Protein linotte
Location in the cell
Cytoplasmic Reliability : 1.071 PlasmaMembrane Reliability : 1.5
Sequence
CDS
ATGGCTGTTCAAAATATTGCGGTGAAATTGTGTGGAATACTTCAGGCAGCAAGTGAGGAGGAAATTACAGCACGAGAGTGGCGTCTTTTAGGACAAGAACAAGATGGTTCACAACTTCTTACTTGGCTTTCAAATACAAAAAATAATGCGGAGGTCCTCAACATTGGCATATATGTAAATAAAACCAAATCTCTGACTATTCTACACACATTTGACAAGAAAACTAATATCATACAAGCTTCAGTGAATGCGACACAAAGTCTTTTAGCATATGTTGTCAAGAAACTTCCAACTGATGATGCTGAGGCAAAAGAGCCTTTGTACTGTGCTTATTTAATGTGTTTACTTCCTGACAAAGAAAGAGTCAGTGAACAAATTGAAGATAGCTCTACAAAACAAATAATGGTACAATATATATACGGGAAGAGTAATAAATACAGCCCTGGTATAAGAAATGACAGATTCCTGCTGTTCAAACATTTGGAATACATAAAAATATATCGCACGCCAATGTTTCTAAATGAATATGACGATGGAACAGAGAAATGGGGCTTTGATGGCATATATCCTGAACCGGAAACCATTGTAAAAGTTTTTTCCTGGGCTCAGTGGGATTGTGTTAATCAGGTTTTGTACCACATTCATTTTCGACAAGCATCGCAGAGCTTCGCTGAGGGCGAGGAGGTGAAATCTCCCTCAGAAATTACGCCCACTCTATCCGCTCTACAGTTCCACGCCGATCTCCCGCACGAGACTGTCGTGAACATCCCCCTAAGTCTGCCTCACGTATCGAGCTCGGCGGAGGGGGCCGCTTGCGGGGCTTACGAGGACGATCCTATACCATTACGAGTCCACGACTGCAGCCTCGACTTGCAGATCGTGTGCGACCAGCGGGGCGTGTTGTGCATCTGTCATCACTACTTGTACAAGCCCCTAGACGATTCCGCCACATCCCTAGTGCTGGAAGACAGCACGGACCTGAAGGACTCCGGAGTCAACTTCGCGTATTCGATAACCCTGCTGCATCACGGCTGCGTGGTGCACACTGTAGTGCCGGACCTGCCGTGGGCCTTGGCCAAGTCGTTACGGCCATCTTACACTTTATACGAAGAAAATCATCTTTTGGTGAATATACCGATGCTGTACACGCATCTGATAGACATCAGCCTCCACCACGAGCCGTGCTGTCACATCGTCCTGCCGGTGGACGAGTGTGACAGCGGGCCCAGCCTCGCGCCCCTGCTGGGCTGGGGTCAGCACGCCATGGTCGACCTCAATACTCTGGACGTGGTGGCCATGGTCATCCCTGAAAAGGATTTGACCGCGACGTTCAAGAGCAACACGGCCATAGAAAATAAACTCGCCATCATACATTACTTGGTGCAGCACGAGAACAATATGGAATTGGCTGAAGAGCTGATATCGTGGATGTGCCTGAATCAGCGCGGCGGGCTGGGCTCGTTCCAGCGCGTCATGCAAGAGTACCTGGTGGCGACCACGTGCGCGGTCACTCGACGCTCGCTACCCTCTTCCGCACTGGGACTTATGAAGATGCTTCCATTCATGCTGCACCTAAAGTTTTCTGATGTCAAGGTTCGCGACATGTGCGTGTCGGTGAGCCAGGAGAAGCTGTGGAGCAGCAGCGCGGTCGTGCTGGCTCCTCGGCAGCGCGTGTGCCAGTTTTCCGAGGACGTGTGGACTAGGGTCTGGGCGTCTCTGTCCGCTCCGTCAGTCACCCGTCTACATCCGACCGTCGTCACCGATAAGCTGCGAGTGAGCCTGCATTGTTACCAGCCGGAAGCGTTGTCTCGGTGCAGCACCCCGCTGTCCCCGAGCGGCCCGGGGCCCGCGCTGGGTTCTCAACGCCGTTCGTCGCTGGGCAACGGGCAGGACCTGCTGCCCTTCTTTGAACTGGACGTGTGCACTGCCAGCAAACAGGAGCATATAGTGGCTGCGGAACTCGCACTAAGCACACTGGTGGCACAGAACTTGCGCGAGGTGAGCGTGCACCTGATGCGCGAGAGCGGCGGCGCGGCGGGCGTGCAGGGCGGCGTGCACGCGGCCGCCACGCGCTGGGCGGCGGCGCAGCTGGATGCCGCGCGCGCGCTGTGCCGGGCGCTGTGCCGCGCGCTCGCCGCCGTGCCCGCGCACCAGCAGCAGCGCGGATTCACTTTCATTGACGATATGGATCCTAACCACGCATACATACTGTTCAAAGTCCTAGAGCAGTACTACCTGGCGACCGACTCCATAGCGTTCCCTGTGCCGCAAGGATTCACATCATTCTTCACGTACTTGGGTTACCGCACTCTGCCCTTTCCCGCCTTCGCGCAGTACGTGCACAGGAGCGTCTTCGAACTGCAGATCGACGTTCTGAAAGCTATCGTGACTGATAAAGAAGAAGACAAGTGCAGCGTGAACAAGAAGCTGCGGCTGATTCAGGTGGCGGGGGAGGGGCGCGGGCGGCGCGCGCTGGCGGCGTGGGGCGCGCGGGCGGCGCTGGTGCTGCGGGCGCGCGACCACGCCGCCGCGCTGCTGTCCGGCGCCGCGCCCGCCCGCCACGACCGCGCGCGACACCGCCCCGTCCGCGACACCGATGATCTAAGAGAAAACATACATTGGAGGAGTGTCAGAACTTAA
Protein
MAVQNIAVKLCGILQAASEEEITAREWRLLGQEQDGSQLLTWLSNTKNNAEVLNIGIYVNKTKSLTILHTFDKKTNIIQASVNATQSLLAYVVKKLPTDDAEAKEPLYCAYLMCLLPDKERVSEQIEDSSTKQIMVQYIYGKSNKYSPGIRNDRFLLFKHLEYIKIYRTPMFLNEYDDGTEKWGFDGIYPEPETIVKVFSWAQWDCVNQVLYHIHFRQASQSFAEGEEVKSPSEITPTLSALQFHADLPHETVVNIPLSLPHVSSSAEGAACGAYEDDPIPLRVHDCSLDLQIVCDQRGVLCICHHYLYKPLDDSATSLVLEDSTDLKDSGVNFAYSITLLHHGCVVHTVVPDLPWALAKSLRPSYTLYEENHLLVNIPMLYTHLIDISLHHEPCCHIVLPVDECDSGPSLAPLLGWGQHAMVDLNTLDVVAMVIPEKDLTATFKSNTAIENKLAIIHYLVQHENNMELAEELISWMCLNQRGGLGSFQRVMQEYLVATTCAVTRRSLPSSALGLMKMLPFMLHLKFSDVKVRDMCVSVSQEKLWSSSAVVLAPRQRVCQFSEDVWTRVWASLSAPSVTRLHPTVVTDKLRVSLHCYQPEALSRCSTPLSPSGPGPALGSQRRSSLGNGQDLLPFFELDVCTASKQEHIVAAELALSTLVAQNLREVSVHLMRESGGAAGVQGGVHAAATRWAAAQLDAARALCRALCRALAAVPAHQQQRGFTFIDDMDPNHAYILFKVLEQYYLATDSIAFPVPQGFTSFFTYLGYRTLPFPAFAQYVHRSVFELQIDVLKAIVTDKEEDKCSVNKKLRLIQVAGEGRGRRALAAWGARAALVLRARDHAAALLSGAAPARHDRARHRPVRDTDDLRENIHWRSVRT
Summary
Miscellaneous
'Linotte' used in the colloquial sentence 'Tete de linotte' literally means 'Scatterbrain' in French.
Similarity
Belongs to the GSAP family.
Keywords
Complete proteome
Reference proteome
Feature
chain Protein pigeon
Uniprot
H9JVD9
A0A2H1W9D1
A0A2W1BH68
A0A194Q4L9
A0A212FI55
A0A1Y1MZ75
+ More
D6W882 A0A1Y1MZ91 A0A2P8Y5Q2 A0A026WSW9 E2B8A2 K7J220 A0A232F4M9 A0A3L8D5Y2 A0A2A3ECT1 A0A151X7I9 A0A195CBF8 A0A154P680 A0A195FQX9 A0A0M9A8R9 A0A0L7RIE2 A0A195BJ88 A0A067QUL2 A0A0A1WR03 A0A0A1X4U6 A0A195DLS8 A0A158NA80 A0A0K8W8R0 A0A0K8UVT5 A0A0K8UG37 E1ZYH0 F4X1F8 A0A034VZE5 A0A034W1Z8 W8BIN6 A0A0L0BV85 A0A1S4EVJ3 A0A336MW29 A0A336L511 A0A182P489 A0A182RK54 A0A182MEC9 A0A1B0CTX2 A0A182JX62 A0A1I8Q6Q5 A0A1A9WHI5 W5JVZ8 A0A2M4D0I3 A0A1B0BES7 A0A182WFG1 A0A1B0G0I0 A0A069DX35 A0A1I8MK75 A0A1I8MK72 A0A1A9XMQ3 A0A1A9VE59 A0A182MZ83 B3MNN3 A0A0P8XES8 Q7Q8H0 A0A182X5W4 A0A182QH41 B0W9W1 A0A0V0G952 A0A182IBJ7 A0A023F1T2 A0A084VL16 A0A1W4UFU7 A0A182U8X0 A0A1W4UUJ1 A0A182VHR7 B4KKF5 A0A1S4GZI6 A0A336MJ49 A0A0Q9X4W9 A0A182KN60 A0A0Q5VLJ6 B3NM80 A0A1Q3F314 B4Q9H9 B4M945 A0A0Q9VYK7 A0A0J9R4G4 B4I5S8 A0A0R1DPI4 B4PAI3 A0A0J9R5G2 A0A182ILI0 B4MTI5 Q24118 M9MRW6 B4JPA0 Q29K33 B4GXF2 A0A0R3NUT2 A0A3B0JTT3 A0A182YN69
D6W882 A0A1Y1MZ91 A0A2P8Y5Q2 A0A026WSW9 E2B8A2 K7J220 A0A232F4M9 A0A3L8D5Y2 A0A2A3ECT1 A0A151X7I9 A0A195CBF8 A0A154P680 A0A195FQX9 A0A0M9A8R9 A0A0L7RIE2 A0A195BJ88 A0A067QUL2 A0A0A1WR03 A0A0A1X4U6 A0A195DLS8 A0A158NA80 A0A0K8W8R0 A0A0K8UVT5 A0A0K8UG37 E1ZYH0 F4X1F8 A0A034VZE5 A0A034W1Z8 W8BIN6 A0A0L0BV85 A0A1S4EVJ3 A0A336MW29 A0A336L511 A0A182P489 A0A182RK54 A0A182MEC9 A0A1B0CTX2 A0A182JX62 A0A1I8Q6Q5 A0A1A9WHI5 W5JVZ8 A0A2M4D0I3 A0A1B0BES7 A0A182WFG1 A0A1B0G0I0 A0A069DX35 A0A1I8MK75 A0A1I8MK72 A0A1A9XMQ3 A0A1A9VE59 A0A182MZ83 B3MNN3 A0A0P8XES8 Q7Q8H0 A0A182X5W4 A0A182QH41 B0W9W1 A0A0V0G952 A0A182IBJ7 A0A023F1T2 A0A084VL16 A0A1W4UFU7 A0A182U8X0 A0A1W4UUJ1 A0A182VHR7 B4KKF5 A0A1S4GZI6 A0A336MJ49 A0A0Q9X4W9 A0A182KN60 A0A0Q5VLJ6 B3NM80 A0A1Q3F314 B4Q9H9 B4M945 A0A0Q9VYK7 A0A0J9R4G4 B4I5S8 A0A0R1DPI4 B4PAI3 A0A0J9R5G2 A0A182ILI0 B4MTI5 Q24118 M9MRW6 B4JPA0 Q29K33 B4GXF2 A0A0R3NUT2 A0A3B0JTT3 A0A182YN69
Pubmed
19121390
28756777
26354079
22118469
28004739
18362917
+ More
19820115 29403074 24508170 20798317 20075255 28648823 30249741 24845553 25830018 21347285 21719571 25348373 24495485 26108605 17510324 20920257 23761445 26334808 25315136 17994087 12364791 25474469 24438588 20966253 22936249 17550304 7576632 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 15632085 25244985
19820115 29403074 24508170 20798317 20075255 28648823 30249741 24845553 25830018 21347285 21719571 25348373 24495485 26108605 17510324 20920257 23761445 26334808 25315136 17994087 12364791 25474469 24438588 20966253 22936249 17550304 7576632 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 15632085 25244985
EMBL
BABH01036029
BABH01036030
BABH01036031
BABH01036032
ODYU01007043
SOQ49452.1
+ More
KZ150213 PZC72150.1 KQ459465 KPJ00294.1 AGBW02008434 OWR53412.1 GEZM01017015 GEZM01017013 JAV90972.1 KQ971307 EFA10899.2 GEZM01017012 GEZM01017011 JAV90973.1 PYGN01000905 PSN39504.1 KK107131 EZA58189.1 GL446286 EFN88104.1 NNAY01000955 OXU25744.1 QOIP01000012 RLU15887.1 KZ288282 PBC29555.1 KQ982446 KYQ56335.1 KQ978009 KYM98137.1 KQ434826 KZC07446.1 KQ981305 KYN42990.1 KQ435726 KOX78253.1 KQ414584 KOC70599.1 KQ976455 KYM85166.1 KK853244 KDR09428.1 GBXI01012808 JAD01484.1 GBXI01007978 JAD06314.1 KQ980734 KYN13850.1 ADTU01010074 GDHF01004778 JAI47536.1 GDHF01021535 GDHF01016228 GDHF01009331 JAI30779.1 JAI36086.1 JAI42983.1 GDHF01026748 GDHF01017070 JAI25566.1 JAI35244.1 GL435215 EFN73767.1 GL888529 EGI59686.1 GAKP01011168 JAC47784.1 GAKP01011164 JAC47788.1 GAMC01009672 GAMC01009670 JAB96883.1 JRES01001284 JRES01000418 KNC23936.1 KNC31450.1 UFQS01002029 UFQT01002029 SSX13007.1 SSX32447.1 SSX13006.1 SSX32446.1 AXCM01002194 AJWK01028078 ADMH02000239 ETN67275.1 GGFL01006430 MBW70608.1 JXJN01013003 CCAG010007529 GBGD01000434 JAC88455.1 CH902620 EDV31120.1 KPU73226.1 AAAB01008944 EAA10179.4 AXCN02002069 DS231866 EDS40528.1 GECL01001695 JAP04429.1 APCN01000708 GBBI01003751 JAC14961.1 ATLV01014396 KE524970 KFB38660.1 CH933807 EDW11605.1 UFQT01001120 SSX28999.1 KRG02805.1 CH954179 KQS61900.1 EDV54680.1 GFDL01013093 JAV21952.1 CM000361 EDX05486.1 CH940654 EDW57721.1 KRF77913.1 CM002910 KMY90963.1 CH480822 EDW55734.1 CM000158 KRJ99148.1 EDW90391.1 KMY90964.1 CH963852 EDW75424.2 U32865 AE014134 AY061465 ADV37093.1 CH916372 EDV98730.1 CH379061 EAL33345.2 CH479195 EDW27263.1 KRT04929.1 OUUW01000010 SPP85515.1
KZ150213 PZC72150.1 KQ459465 KPJ00294.1 AGBW02008434 OWR53412.1 GEZM01017015 GEZM01017013 JAV90972.1 KQ971307 EFA10899.2 GEZM01017012 GEZM01017011 JAV90973.1 PYGN01000905 PSN39504.1 KK107131 EZA58189.1 GL446286 EFN88104.1 NNAY01000955 OXU25744.1 QOIP01000012 RLU15887.1 KZ288282 PBC29555.1 KQ982446 KYQ56335.1 KQ978009 KYM98137.1 KQ434826 KZC07446.1 KQ981305 KYN42990.1 KQ435726 KOX78253.1 KQ414584 KOC70599.1 KQ976455 KYM85166.1 KK853244 KDR09428.1 GBXI01012808 JAD01484.1 GBXI01007978 JAD06314.1 KQ980734 KYN13850.1 ADTU01010074 GDHF01004778 JAI47536.1 GDHF01021535 GDHF01016228 GDHF01009331 JAI30779.1 JAI36086.1 JAI42983.1 GDHF01026748 GDHF01017070 JAI25566.1 JAI35244.1 GL435215 EFN73767.1 GL888529 EGI59686.1 GAKP01011168 JAC47784.1 GAKP01011164 JAC47788.1 GAMC01009672 GAMC01009670 JAB96883.1 JRES01001284 JRES01000418 KNC23936.1 KNC31450.1 UFQS01002029 UFQT01002029 SSX13007.1 SSX32447.1 SSX13006.1 SSX32446.1 AXCM01002194 AJWK01028078 ADMH02000239 ETN67275.1 GGFL01006430 MBW70608.1 JXJN01013003 CCAG010007529 GBGD01000434 JAC88455.1 CH902620 EDV31120.1 KPU73226.1 AAAB01008944 EAA10179.4 AXCN02002069 DS231866 EDS40528.1 GECL01001695 JAP04429.1 APCN01000708 GBBI01003751 JAC14961.1 ATLV01014396 KE524970 KFB38660.1 CH933807 EDW11605.1 UFQT01001120 SSX28999.1 KRG02805.1 CH954179 KQS61900.1 EDV54680.1 GFDL01013093 JAV21952.1 CM000361 EDX05486.1 CH940654 EDW57721.1 KRF77913.1 CM002910 KMY90963.1 CH480822 EDW55734.1 CM000158 KRJ99148.1 EDW90391.1 KMY90964.1 CH963852 EDW75424.2 U32865 AE014134 AY061465 ADV37093.1 CH916372 EDV98730.1 CH379061 EAL33345.2 CH479195 EDW27263.1 KRT04929.1 OUUW01000010 SPP85515.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000007266
UP000245037
UP000053097
+ More
UP000008237 UP000002358 UP000215335 UP000279307 UP000242457 UP000075809 UP000078542 UP000076502 UP000078541 UP000053105 UP000053825 UP000078540 UP000027135 UP000078492 UP000005205 UP000000311 UP000007755 UP000037069 UP000075885 UP000075900 UP000075883 UP000092461 UP000075881 UP000095300 UP000091820 UP000000673 UP000092460 UP000075920 UP000092444 UP000095301 UP000092443 UP000078200 UP000075884 UP000007801 UP000007062 UP000076407 UP000075886 UP000002320 UP000075840 UP000030765 UP000192221 UP000075902 UP000075903 UP000009192 UP000075882 UP000008711 UP000000304 UP000008792 UP000001292 UP000002282 UP000075880 UP000007798 UP000000803 UP000001070 UP000001819 UP000008744 UP000268350 UP000076408
UP000008237 UP000002358 UP000215335 UP000279307 UP000242457 UP000075809 UP000078542 UP000076502 UP000078541 UP000053105 UP000053825 UP000078540 UP000027135 UP000078492 UP000005205 UP000000311 UP000007755 UP000037069 UP000075885 UP000075900 UP000075883 UP000092461 UP000075881 UP000095300 UP000091820 UP000000673 UP000092460 UP000075920 UP000092444 UP000095301 UP000092443 UP000078200 UP000075884 UP000007801 UP000007062 UP000076407 UP000075886 UP000002320 UP000075840 UP000030765 UP000192221 UP000075902 UP000075903 UP000009192 UP000075882 UP000008711 UP000000304 UP000008792 UP000001292 UP000002282 UP000075880 UP000007798 UP000000803 UP000001070 UP000001819 UP000008744 UP000268350 UP000076408
Pfam
PF14959 GSAP-16
ProteinModelPortal
H9JVD9
A0A2H1W9D1
A0A2W1BH68
A0A194Q4L9
A0A212FI55
A0A1Y1MZ75
+ More
D6W882 A0A1Y1MZ91 A0A2P8Y5Q2 A0A026WSW9 E2B8A2 K7J220 A0A232F4M9 A0A3L8D5Y2 A0A2A3ECT1 A0A151X7I9 A0A195CBF8 A0A154P680 A0A195FQX9 A0A0M9A8R9 A0A0L7RIE2 A0A195BJ88 A0A067QUL2 A0A0A1WR03 A0A0A1X4U6 A0A195DLS8 A0A158NA80 A0A0K8W8R0 A0A0K8UVT5 A0A0K8UG37 E1ZYH0 F4X1F8 A0A034VZE5 A0A034W1Z8 W8BIN6 A0A0L0BV85 A0A1S4EVJ3 A0A336MW29 A0A336L511 A0A182P489 A0A182RK54 A0A182MEC9 A0A1B0CTX2 A0A182JX62 A0A1I8Q6Q5 A0A1A9WHI5 W5JVZ8 A0A2M4D0I3 A0A1B0BES7 A0A182WFG1 A0A1B0G0I0 A0A069DX35 A0A1I8MK75 A0A1I8MK72 A0A1A9XMQ3 A0A1A9VE59 A0A182MZ83 B3MNN3 A0A0P8XES8 Q7Q8H0 A0A182X5W4 A0A182QH41 B0W9W1 A0A0V0G952 A0A182IBJ7 A0A023F1T2 A0A084VL16 A0A1W4UFU7 A0A182U8X0 A0A1W4UUJ1 A0A182VHR7 B4KKF5 A0A1S4GZI6 A0A336MJ49 A0A0Q9X4W9 A0A182KN60 A0A0Q5VLJ6 B3NM80 A0A1Q3F314 B4Q9H9 B4M945 A0A0Q9VYK7 A0A0J9R4G4 B4I5S8 A0A0R1DPI4 B4PAI3 A0A0J9R5G2 A0A182ILI0 B4MTI5 Q24118 M9MRW6 B4JPA0 Q29K33 B4GXF2 A0A0R3NUT2 A0A3B0JTT3 A0A182YN69
D6W882 A0A1Y1MZ91 A0A2P8Y5Q2 A0A026WSW9 E2B8A2 K7J220 A0A232F4M9 A0A3L8D5Y2 A0A2A3ECT1 A0A151X7I9 A0A195CBF8 A0A154P680 A0A195FQX9 A0A0M9A8R9 A0A0L7RIE2 A0A195BJ88 A0A067QUL2 A0A0A1WR03 A0A0A1X4U6 A0A195DLS8 A0A158NA80 A0A0K8W8R0 A0A0K8UVT5 A0A0K8UG37 E1ZYH0 F4X1F8 A0A034VZE5 A0A034W1Z8 W8BIN6 A0A0L0BV85 A0A1S4EVJ3 A0A336MW29 A0A336L511 A0A182P489 A0A182RK54 A0A182MEC9 A0A1B0CTX2 A0A182JX62 A0A1I8Q6Q5 A0A1A9WHI5 W5JVZ8 A0A2M4D0I3 A0A1B0BES7 A0A182WFG1 A0A1B0G0I0 A0A069DX35 A0A1I8MK75 A0A1I8MK72 A0A1A9XMQ3 A0A1A9VE59 A0A182MZ83 B3MNN3 A0A0P8XES8 Q7Q8H0 A0A182X5W4 A0A182QH41 B0W9W1 A0A0V0G952 A0A182IBJ7 A0A023F1T2 A0A084VL16 A0A1W4UFU7 A0A182U8X0 A0A1W4UUJ1 A0A182VHR7 B4KKF5 A0A1S4GZI6 A0A336MJ49 A0A0Q9X4W9 A0A182KN60 A0A0Q5VLJ6 B3NM80 A0A1Q3F314 B4Q9H9 B4M945 A0A0Q9VYK7 A0A0J9R4G4 B4I5S8 A0A0R1DPI4 B4PAI3 A0A0J9R5G2 A0A182ILI0 B4MTI5 Q24118 M9MRW6 B4JPA0 Q29K33 B4GXF2 A0A0R3NUT2 A0A3B0JTT3 A0A182YN69
Ontologies
GO
PANTHER
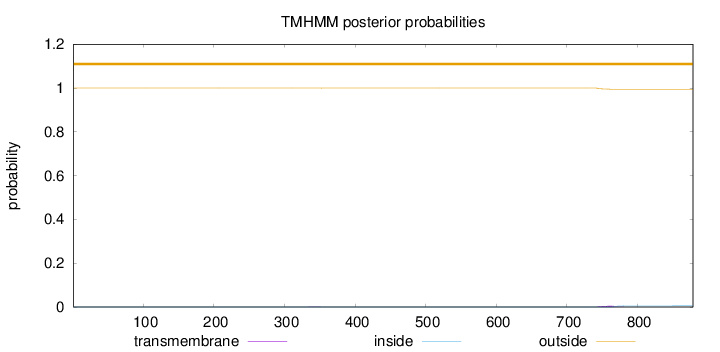
Topology
Length:
879
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11933
Exp number, first 60 AAs:
0.00059
Total prob of N-in:
0.00033
outside
1 - 879
Population Genetic Test Statistics
Pi
257.795788
Theta
197.774166
Tajima's D
0.740457
CLR
0.381384
CSRT
0.578121093945303
Interpretation
Uncertain
