Gene
KWMTBOMO02439
Pre Gene Modal
BGIBMGA013501
Annotation
PREDICTED:_structural_maintenance_of_chromosomes_protein_6_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.519 Nuclear Reliability : 1.81
Sequence
CDS
ATGCGTAACTGGTTTTGTCTCGAAAATTTGAAAATAAAGTTTAACAGAAACGTAAATTTTATCGTTGGTCGTAACGGATGTGGAAAAAGTGCTATTCTATCTGCATTAGTGATTGGCTTGGGCAGCAGAGCTGATATAACTTACAGAGGAAATAATGTGCAATCTTTAATCAGGCGTGGAGCAAATTCAGCAACCATTGAGATTCAACTGAGAAACAGTAGTTCTAGAGCATACAAACGTCACATTTATGGCGATCACATTACAATAATCAGAGAAATCAATGCAAGTGGTTCGCATTATAAAATTAAAGCTGCATCAGGTGAAATAATATCAACTAGGTTAAAAGAAGTTGAAGCAATAAAACATGCTCTTGGAATACAAGTTGATAATCCCATTTCGGTGCTTAATCAAGATGATGCTAGGAGTTTCCTTGGTAAAGGACCTGAGAAGATGTATGCCCTGTTCCGGAAAGCCACAAATTTGGACCAGGCAGAAGATAAATATAAAGAAACATTGAAAACATGTGATGAGGCAATTGAACATTTAAAAATGAAAGAGCAAGCTTCGAAAGAGTTGGAAAAAGAATACCATAGTCTAAGGGCGAGCTATGATCAGATGAGGTCATACGAAGAATTTGAGCAGCAAATTCAGCAATTACAAAATGAATATTACTGGAGAGAGGTGGTTGAGGTTGAGACAGAAGCACATGATGCTGAGACACAGTACAATACTGTTTGTAAGAAGATCACTGAGCTCACACAGAAGGGTCAAGACATTGAACAAATACATAGAACAGGATTGAGTGCTCTTGAAGTCTTAAAGAGAGAGCGAGATGAAAAATCGTTACAAAAGAATGCCTTACAACAAGATGTGTCGACGTACGAATCTCAAGCCCGAGAAGCTCAGAGTTCGTTGAGAACGGTGCAACAAACCGTTCAGAAATACCAGGACAGGTTCGACAGAGAACAGAAGAAAGTCTTAGATTTACAGAGGGAGATAGGGAATATACAATCGATTAACCCAACAACTGCGCGACCTGGAGTCCTGCTCCAGCGACTCGCTGGCCGTGTACGGCAACAGCATGGTGACGCTCCGGAAGGAGCTCTGCGCCGCCTGCCGCAGGTGTACGGGCCCGCGCGCAAGCCGGCCGTCTCGTGCAGCGCGTTCCTGCGGCGGCGCCACGACGTGCGGGCGCGCGAGGTGCGCGGGGCGGGGGGCGGCGCGGCGGGGGGCGCCCCCCTGCGCTCGGCGCTGGGCTGGCTGCAGGTCGACGACCCCGAGGTCTACAACCACCTCGTGGACGCGCTGCGCCTGGAGACCGTGTTGCTTGTACCCGATCACCAGACAGCGGTGGAGCTGTGCTCGGAGGAGCACCGCGTGCCGCGCCACCTCAGCAAGGCCGTGAGCCGCGACTGCGCGGAGTACTACCCCGCCCCGCGCTACCGCTCCTACGGCGTGCGCGCCCGCCCCCCGCGCTACCTGCAGCACTCCTCGCGCGACCGCACCAGGCAGCTAAAGTCGGAGATACAAGAAGCGCAAGTAAAGTTGCGAGAGCTGGAGGGCGGCGCCCGCGCGTGCGAGCAGGAGGCGGAGCGCGCCCGCCAGTACGAGCGCGCGCTCGCCGACACGCTGCACGCGCTCGCGCTCGACAAGATCGACAAAGAGGAGAAGGAGAAGATCGCCATAGCTGAACTCGAGCAGTACCAGGAGCCGCGACAAACTGTGCTGGAGGACGAATTGAAATTGTCCAAAGAAAAAGCCCAAAGCGTACAAGAACAGTTGAACGCGCAGTTAGCGGAAGAGCAGCGTTGGAGAGGCCAGCACCAGCAGCTCGATAACAAAGTGAGGGAGCTCCGGAACCGACTGTCGGACCTCACGTCCGCCTGCAGGGACCTCACGGAAGAAATAGATCGGGAGCAAATCAAATTAGAGAATATTGCGTCTCAAATAACCGAATGTGAAGTTCGCGTGCAAAAATACCGCAAGGAGTTGTCTCGAACCAAAGCAGCCTTGGAAGAGAAAAAGGAAGTCGTCAGAACAAAAATTGAAGAAACGTCGAAGCTTTGTCCTCGTGTTCGCAACCCCAGACAAAAAGCATTGATAACGGAAGAATTGAAAAATACAAAAGATAAGATGAACGCGATGCGATGCAACGGCATGACGAAGGTGCAGGTGGCCGAGAAACTGATCGTCGTTAAGAAGAAATATCATGAGATTAGCGCGGAACTGCAGCAGCTCGGAGCCGTCATTGAATCCATCAAGGAGACCGCCAAGCAACATCTGCAGTTCAGTCACTTCGCGCAGACCTACATCATCAGGCAGCTGTACTACATATTCCAGTCCATCCTGACGCTGAGAGGATACTGCGTGAGTTCCTGCTTGTTCCCTTATGACTACTAA
Protein
MRNWFCLENLKIKFNRNVNFIVGRNGCGKSAILSALVIGLGSRADITYRGNNVQSLIRRGANSATIEIQLRNSSSRAYKRHIYGDHITIIREINASGSHYKIKAASGEIISTRLKEVEAIKHALGIQVDNPISVLNQDDARSFLGKGPEKMYALFRKATNLDQAEDKYKETLKTCDEAIEHLKMKEQASKELEKEYHSLRASYDQMRSYEEFEQQIQQLQNEYYWREVVEVETEAHDAETQYNTVCKKITELTQKGQDIEQIHRTGLSALEVLKRERDEKSLQKNALQQDVSTYESQAREAQSSLRTVQQTVQKYQDRFDREQKKVLDLQREIGNIQSINPTTARPGVLLQRLAGRVRQQHGDAPEGALRRLPQVYGPARKPAVSCSAFLRRRHDVRAREVRGAGGGAAGGAPLRSALGWLQVDDPEVYNHLVDALRLETVLLVPDHQTAVELCSEEHRVPRHLSKAVSRDCAEYYPAPRYRSYGVRARPPRYLQHSSRDRTRQLKSEIQEAQVKLRELEGGARACEQEAERARQYERALADTLHALALDKIDKEEKEKIAIAELEQYQEPRQTVLEDELKLSKEKAQSVQEQLNAQLAEEQRWRGQHQQLDNKVRELRNRLSDLTSACRDLTEEIDREQIKLENIASQITECEVRVQKYRKELSRTKAALEEKKEVVRTKIEETSKLCPRVRNPRQKALITEELKNTKDKMNAMRCNGMTKVQVAEKLIVVKKKYHEISAELQQLGAVIESIKETAKQHLQFSHFAQTYIIRQLYYIFQSILTLRGYCVSSCLFPYDY
Summary
Uniprot
EMBL
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
PDB
5F3W
E-value=6.51167e-06,
Score=122
Ontologies
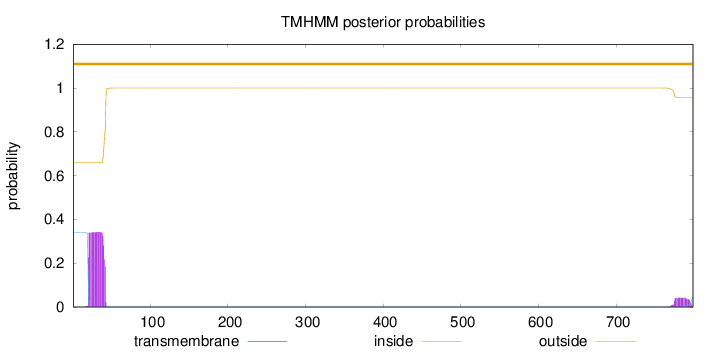
Topology
Length:
799
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.32183999999999
Exp number, first 60 AAs:
7.4083
Total prob of N-in:
0.34153
outside
1 - 799
Population Genetic Test Statistics
Pi
135.669968
Theta
135.967582
Tajima's D
0.097359
CLR
0.275113
CSRT
0.392180390980451
Interpretation
Uncertain
