Gene
KWMTBOMO02432 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013498
Annotation
PREDICTED:_dolichyl-diphosphooligosaccharide--protein_glycosyltransferase_subunit_2_[Plutella_xylostella]
Full name
XK-related protein
Location in the cell
Peroxisomal Reliability : 1.07
Sequence
CDS
ATGTACTTTCGAGTACAATTGCTCATCTTAGTGAGTATAGCAGTAAGTAATGCCAACATTCTCCAAGGCACTGTGGATATCAATCGCCTTCAAACCATACTTAAAGACAACTTAAAGAGCAAAGATGTGGGCACCCTGTATTATGCTGTTCGAGGATTGAAACAGTTGAAAGCTGATGTGCCAAATATTTGTGAGGACCTCAAAACGATAAAGTATGATGTCAAGAACTTAGAGCAAGTGTTTTACCTCACAAACTTGGCCCTTCTTACAAACTGTCAGAATTCACTCAAGCCTGAAGTATTGGCGACTCCAACCCAAGCACTAGACAAAAAGGATGTCACCATACAGGAACTGTACTTAGCTGTGTATACACTAAAAGCTCTTGGTAAAGGAACTATTTATGATAAAGAGGATGCTTTAAAAAATCTTATTCAGCTGCTGAAAAAGGATGACACACCTGCCAATTATGGTTATGTATTTGCATTATGTGAGCACATGGGCTGTGGGCTATGGACAACTTCGCATGCTGAAGGAGTTATACTTGCTGCTGATGAGACAGACTCTCGTGCCTTACATTTTGAAGGAGGCCTACCTGTCACATCACAACTTTTGACTACAATAACCAGGACATACAAAGCTCAAAAGAAGCCGACGCCATTCACGGAGGAGCAACGGTACAAGTTCGCCGAGTACCTGTTGTCGCGTCGGTCCGTCAACACCGCCAAGGGCGTGACGCTCCTCCTGGAGGCAGCCACTGCCATCGCGGACGATGAACCCACACCGATCAGCATTGCGGTCAAAGGCAAGAAGTATGTGACCTCTGAATCTGACAGCATCGAGTTCACCATCACCGACCTCATCGGCAGGCCGGTCCGCAACCTCAAACCTGAAGAAGTAATCGCACAGTCTGGCACCAGGCTCGCCGACGACGTCGTTGTGCTATCCAAACAGCCTTTAACCCAGAAACCTAACGAACCGACTTTCGTCCTTAATCTAAACAAGATTAAATCACAATATGGCGTGTACAAGATCGCTTTAAGTGCCGGGACGAAGACGACCAGCGTGAACGTGGCCGTGCTGGGAGAGATCCAGGTCGCCAATGTCGAGGTCGGCGTCGGCGACGTGGACGGCACCACCAGCCCTAAGATTACAACCATCACTTACCCGAATAAACTGGTGGATAAGTTGCTGGCTGATCATATGCAGAAAGTGAGCCTGAAGTTCACAGTTCGCGACAAATGGAACAAGCCCGTTTTAGTTCAACAAGCTTTTGTAAGGGTTGCCAGCACAGAGACTGACGATGAAACTATCTTCGTTGCAGAACCTGACAACACTAAAGCATATAAAGTTGAATTGAACATCGGCAGCATAGCGAAGCACCTCAACTACGCGTCGGGCTCGTACTCCCTGAGCGTGTACGTAGGAGACAGCGCCGTGTCCAGCCCCGTAGCCTGGCTGCTGGGAGACGTCTCCTTCAACTTCGGCAAGGACGCCAATGCTGTTGGGGTGGCGCGGGCGACGGGTGCGGCGGGGGGGCGGCTGCCGGAGCTGGCGCACACGTTCCGCGCGGCGGAGGCGCGGCCGGCGCGCGCGCTGTCGGACGTGTGCAGCGCGCTGTGCGCGGCGCCGCTGCTGGTGCTCCTGCTGCTGTGGGCGCGGATCGGCATCAACTTGCGGAACTTCCCCCTCGTACCCAGCGCGCTTATATTCCATCTAGCCCTTGGAGCATCTCTCGGGCTGTACGTCGTGCTGTGGCTCCAGCTGACAATGTTCGAAGCCATCCGCTACCTGCTGCCCCTGGGCGCCGTCACCTTCCTCTCCGGACATCGCCTGCTGCAGCGTCTCGTGAAAGACAAAACCTCCAAATAA
Protein
MYFRVQLLILVSIAVSNANILQGTVDINRLQTILKDNLKSKDVGTLYYAVRGLKQLKADVPNICEDLKTIKYDVKNLEQVFYLTNLALLTNCQNSLKPEVLATPTQALDKKDVTIQELYLAVYTLKALGKGTIYDKEDALKNLIQLLKKDDTPANYGYVFALCEHMGCGLWTTSHAEGVILAADETDSRALHFEGGLPVTSQLLTTITRTYKAQKKPTPFTEEQRYKFAEYLLSRRSVNTAKGVTLLLEAATAIADDEPTPISIAVKGKKYVTSESDSIEFTITDLIGRPVRNLKPEEVIAQSGTRLADDVVVLSKQPLTQKPNEPTFVLNLNKIKSQYGVYKIALSAGTKTTSVNVAVLGEIQVANVEVGVGDVDGTTSPKITTITYPNKLVDKLLADHMQKVSLKFTVRDKWNKPVLVQQAFVRVASTETDDETIFVAEPDNTKAYKVELNIGSIAKHLNYASGSYSLSVYVGDSAVSSPVAWLLGDVSFNFGKDANAVGVARATGAAGGRLPELAHTFRAAEARPARALSDVCSALCAAPLLVLLLLWARIGINLRNFPLVPSALIFHLALGASLGLYVVLWLQLTMFEAIRYLLPLGAVTFLSGHRLLQRLVKDKTSK
Summary
Similarity
Belongs to the XK family.
Uniprot
H9JVD5
A0A2W1BAP9
A0A2H1W5E4
A0A1E1WVG2
A0A0N1IDA9
S4PMW5
+ More
A0A2A4JMM6 A0A194Q431 A0A212FIQ3 A0A0L7LTH0 A0A0M3QVG2 B4LKS1 A0A0M8ZNE6 A0A2A3E236 B4KQV8 K7JAD4 B4J9J3 A0A0C9QA03 B4P536 A0A1W4VDV6 A0A0K8TU52 A0A1L8E194 A0A1L8E1D4 A0A088A702 A0A0B4LFR4 A0A131XNK8 Q7K110 B4QBQ8 B4HN77 A0A232FBP6 A0A0L7RIL1 B3NLK0 B3MIC3 A0A067QE65 A0A2J7QDE8 A0A182NHK5 A0A154NZJ3 B0VZR0 A0A1L8EBK6 A0A1L8EC07 A0A1L8EBM7 A0A3B0J1P4 A0A1B0CEI4 A0A1I8NVP8 A0A1L8EBN3 T1PCA4 A0A1I8NG54 A0A1I8NG52 A0A023FDH8 A0A0J7LBH9 B5E0Y7 A0A1L8EBK7 B4GG23 A0A023GLS6 G3MM45 A0A023GNM8 A0A182MWC5 E2BN86 A0A023FWW6 A0A131XHF6 A0A0L0CEA5 A0A182I3Q6 B4NNG6 A0A182WIS6 A0A084VFB9 A0A182JNY7 A0A034WFW4 A0A182X6F3 A0A131Z110 A0A182P3I7 Q7QJS7 U5EVZ5 A0A182KRU4 A0A182QE28 A0A224Z4M0 A0A182HB54 A0A1W7RAP6 A0A0K8WKN7 A0A0K8UY29 A0A1A9UIX8 A0A1B0FMY9 E0W4C5 A0A1B6CY97 L7MII0 A0A1B6DAG0 D3TPY1 A0A026WLG8 A0A2M4BH05 A0A195CLC7 A0A2M4BHQ8 A0A182FIA1 A0A182J203 A0A1B0A9B8 A0A2M4BH22 A0A182VBE3 A0A1B0AKJ8 A0A0A1WGB5 A0A182TTD8 A0A1L8E9L3 A0A151XHZ1
A0A2A4JMM6 A0A194Q431 A0A212FIQ3 A0A0L7LTH0 A0A0M3QVG2 B4LKS1 A0A0M8ZNE6 A0A2A3E236 B4KQV8 K7JAD4 B4J9J3 A0A0C9QA03 B4P536 A0A1W4VDV6 A0A0K8TU52 A0A1L8E194 A0A1L8E1D4 A0A088A702 A0A0B4LFR4 A0A131XNK8 Q7K110 B4QBQ8 B4HN77 A0A232FBP6 A0A0L7RIL1 B3NLK0 B3MIC3 A0A067QE65 A0A2J7QDE8 A0A182NHK5 A0A154NZJ3 B0VZR0 A0A1L8EBK6 A0A1L8EC07 A0A1L8EBM7 A0A3B0J1P4 A0A1B0CEI4 A0A1I8NVP8 A0A1L8EBN3 T1PCA4 A0A1I8NG54 A0A1I8NG52 A0A023FDH8 A0A0J7LBH9 B5E0Y7 A0A1L8EBK7 B4GG23 A0A023GLS6 G3MM45 A0A023GNM8 A0A182MWC5 E2BN86 A0A023FWW6 A0A131XHF6 A0A0L0CEA5 A0A182I3Q6 B4NNG6 A0A182WIS6 A0A084VFB9 A0A182JNY7 A0A034WFW4 A0A182X6F3 A0A131Z110 A0A182P3I7 Q7QJS7 U5EVZ5 A0A182KRU4 A0A182QE28 A0A224Z4M0 A0A182HB54 A0A1W7RAP6 A0A0K8WKN7 A0A0K8UY29 A0A1A9UIX8 A0A1B0FMY9 E0W4C5 A0A1B6CY97 L7MII0 A0A1B6DAG0 D3TPY1 A0A026WLG8 A0A2M4BH05 A0A195CLC7 A0A2M4BHQ8 A0A182FIA1 A0A182J203 A0A1B0A9B8 A0A2M4BH22 A0A182VBE3 A0A1B0AKJ8 A0A0A1WGB5 A0A182TTD8 A0A1L8E9L3 A0A151XHZ1
Pubmed
19121390
28756777
26354079
23622113
22118469
26227816
+ More
17994087 20075255 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 28648823 24845553 25315136 15632085 22216098 20798317 28049606 26108605 24438588 25348373 26830274 12364791 14747013 17210077 20966253 28797301 26483478 20566863 25576852 20353571 24508170 25830018
17994087 20075255 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 28648823 24845553 25315136 15632085 22216098 20798317 28049606 26108605 24438588 25348373 26830274 12364791 14747013 17210077 20966253 28797301 26483478 20566863 25576852 20353571 24508170 25830018
EMBL
BABH01036046
KZ150213
PZC72158.1
ODYU01006430
SOQ48268.1
GDQN01010411
+ More
GDQN01000258 JAT80643.1 JAT90796.1 KQ460796 KPJ12175.1 GAIX01003330 JAA89230.1 NWSH01001095 PCG72650.1 KQ459465 KPJ00302.1 AGBW02008342 OWR53618.1 JTDY01000137 KOB78654.1 CP012524 ALC42323.1 CH940648 EDW61794.1 KQ435969 KOX67830.1 KZ288432 PBC25803.1 CH933808 EDW09307.1 AAZX01005687 CH916367 EDW02500.1 GBYB01011233 JAG81000.1 CM000158 EDW91737.1 GDAI01000123 JAI17480.1 GFDF01001584 JAV12500.1 GFDF01001585 JAV12499.1 AE013599 AHN56345.1 GEFM01007013 JAP68783.1 AY069461 AAF57793.1 AAL39606.1 CM000362 CM002911 EDX07568.1 KMY94679.1 CH480816 EDW48359.1 NNAY01000481 OXU28075.1 KQ414583 KOC70802.1 CH954179 EDV54975.1 CH902619 EDV36971.1 KK853953 KDQ71487.1 NEVH01015824 PNF26612.1 KQ434786 KZC05031.1 DS231815 EDS35152.1 GFDG01002659 JAV16140.1 GFDG01002657 JAV16142.1 GFDG01002661 JAV16138.1 OUUW01000001 SPP74907.1 AJWK01008901 GFDG01002658 JAV16141.1 KA645568 AFP60197.1 GBBK01004551 JAC19931.1 LBMM01000031 KMR05267.1 CM000071 EDY69429.1 GFDG01002662 JAV16137.1 CH479183 EDW35443.1 GBBM01000629 JAC34789.1 JO842946 AEO34563.1 GBBM01000630 JAC34788.1 AXCM01000880 GL449385 EFN82848.1 GBBL01000964 JAC26356.1 GEFH01003470 JAP65111.1 JRES01000505 KNC30537.1 APCN01000217 CH964282 EDW85905.1 ATLV01012374 KE524785 KFB36663.1 GAKP01004481 JAC54471.1 GEDV01004411 JAP84146.1 AAAB01008807 EAA04099.4 GANO01000780 JAB59091.1 AXCN02000593 GFPF01010357 MAA21503.1 JXUM01124142 KQ566796 KXJ69818.1 GFAH01000177 JAV48212.1 GDHF01000884 JAI51430.1 GDHF01020866 JAI31448.1 CCAG010011323 DS235886 EEB20481.1 GEDC01018848 JAS18450.1 GACK01001013 JAA64021.1 GEDC01014723 JAS22575.1 EZ423483 ADD19759.1 KK107182 EZA55944.1 GGFJ01003195 MBW52336.1 KQ977642 KYN00889.1 GGFJ01003197 MBW52338.1 GGFJ01003196 MBW52337.1 JXJN01025954 GBXI01016849 JAC97442.1 GFDG01003415 JAV15384.1 KQ982101 KYQ60019.1
GDQN01000258 JAT80643.1 JAT90796.1 KQ460796 KPJ12175.1 GAIX01003330 JAA89230.1 NWSH01001095 PCG72650.1 KQ459465 KPJ00302.1 AGBW02008342 OWR53618.1 JTDY01000137 KOB78654.1 CP012524 ALC42323.1 CH940648 EDW61794.1 KQ435969 KOX67830.1 KZ288432 PBC25803.1 CH933808 EDW09307.1 AAZX01005687 CH916367 EDW02500.1 GBYB01011233 JAG81000.1 CM000158 EDW91737.1 GDAI01000123 JAI17480.1 GFDF01001584 JAV12500.1 GFDF01001585 JAV12499.1 AE013599 AHN56345.1 GEFM01007013 JAP68783.1 AY069461 AAF57793.1 AAL39606.1 CM000362 CM002911 EDX07568.1 KMY94679.1 CH480816 EDW48359.1 NNAY01000481 OXU28075.1 KQ414583 KOC70802.1 CH954179 EDV54975.1 CH902619 EDV36971.1 KK853953 KDQ71487.1 NEVH01015824 PNF26612.1 KQ434786 KZC05031.1 DS231815 EDS35152.1 GFDG01002659 JAV16140.1 GFDG01002657 JAV16142.1 GFDG01002661 JAV16138.1 OUUW01000001 SPP74907.1 AJWK01008901 GFDG01002658 JAV16141.1 KA645568 AFP60197.1 GBBK01004551 JAC19931.1 LBMM01000031 KMR05267.1 CM000071 EDY69429.1 GFDG01002662 JAV16137.1 CH479183 EDW35443.1 GBBM01000629 JAC34789.1 JO842946 AEO34563.1 GBBM01000630 JAC34788.1 AXCM01000880 GL449385 EFN82848.1 GBBL01000964 JAC26356.1 GEFH01003470 JAP65111.1 JRES01000505 KNC30537.1 APCN01000217 CH964282 EDW85905.1 ATLV01012374 KE524785 KFB36663.1 GAKP01004481 JAC54471.1 GEDV01004411 JAP84146.1 AAAB01008807 EAA04099.4 GANO01000780 JAB59091.1 AXCN02000593 GFPF01010357 MAA21503.1 JXUM01124142 KQ566796 KXJ69818.1 GFAH01000177 JAV48212.1 GDHF01000884 JAI51430.1 GDHF01020866 JAI31448.1 CCAG010011323 DS235886 EEB20481.1 GEDC01018848 JAS18450.1 GACK01001013 JAA64021.1 GEDC01014723 JAS22575.1 EZ423483 ADD19759.1 KK107182 EZA55944.1 GGFJ01003195 MBW52336.1 KQ977642 KYN00889.1 GGFJ01003197 MBW52338.1 GGFJ01003196 MBW52337.1 JXJN01025954 GBXI01016849 JAC97442.1 GFDG01003415 JAV15384.1 KQ982101 KYQ60019.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000037510
+ More
UP000092553 UP000008792 UP000053105 UP000242457 UP000009192 UP000002358 UP000001070 UP000002282 UP000192221 UP000005203 UP000000803 UP000000304 UP000001292 UP000215335 UP000053825 UP000008711 UP000007801 UP000027135 UP000235965 UP000075884 UP000076502 UP000002320 UP000268350 UP000092461 UP000095300 UP000095301 UP000036403 UP000001819 UP000008744 UP000075883 UP000008237 UP000037069 UP000075840 UP000007798 UP000075920 UP000030765 UP000075881 UP000076407 UP000075885 UP000007062 UP000075882 UP000075886 UP000069940 UP000249989 UP000078200 UP000092444 UP000009046 UP000053097 UP000078542 UP000069272 UP000075880 UP000092445 UP000075903 UP000092460 UP000075902 UP000075809
UP000092553 UP000008792 UP000053105 UP000242457 UP000009192 UP000002358 UP000001070 UP000002282 UP000192221 UP000005203 UP000000803 UP000000304 UP000001292 UP000215335 UP000053825 UP000008711 UP000007801 UP000027135 UP000235965 UP000075884 UP000076502 UP000002320 UP000268350 UP000092461 UP000095300 UP000095301 UP000036403 UP000001819 UP000008744 UP000075883 UP000008237 UP000037069 UP000075840 UP000007798 UP000075920 UP000030765 UP000075881 UP000076407 UP000075885 UP000007062 UP000075882 UP000075886 UP000069940 UP000249989 UP000078200 UP000092444 UP000009046 UP000053097 UP000078542 UP000069272 UP000075880 UP000092445 UP000075903 UP000092460 UP000075902 UP000075809
Pfam
Interpro
SUPFAM
SSF48464
SSF48464
Gene 3D
CDD
ProteinModelPortal
H9JVD5
A0A2W1BAP9
A0A2H1W5E4
A0A1E1WVG2
A0A0N1IDA9
S4PMW5
+ More
A0A2A4JMM6 A0A194Q431 A0A212FIQ3 A0A0L7LTH0 A0A0M3QVG2 B4LKS1 A0A0M8ZNE6 A0A2A3E236 B4KQV8 K7JAD4 B4J9J3 A0A0C9QA03 B4P536 A0A1W4VDV6 A0A0K8TU52 A0A1L8E194 A0A1L8E1D4 A0A088A702 A0A0B4LFR4 A0A131XNK8 Q7K110 B4QBQ8 B4HN77 A0A232FBP6 A0A0L7RIL1 B3NLK0 B3MIC3 A0A067QE65 A0A2J7QDE8 A0A182NHK5 A0A154NZJ3 B0VZR0 A0A1L8EBK6 A0A1L8EC07 A0A1L8EBM7 A0A3B0J1P4 A0A1B0CEI4 A0A1I8NVP8 A0A1L8EBN3 T1PCA4 A0A1I8NG54 A0A1I8NG52 A0A023FDH8 A0A0J7LBH9 B5E0Y7 A0A1L8EBK7 B4GG23 A0A023GLS6 G3MM45 A0A023GNM8 A0A182MWC5 E2BN86 A0A023FWW6 A0A131XHF6 A0A0L0CEA5 A0A182I3Q6 B4NNG6 A0A182WIS6 A0A084VFB9 A0A182JNY7 A0A034WFW4 A0A182X6F3 A0A131Z110 A0A182P3I7 Q7QJS7 U5EVZ5 A0A182KRU4 A0A182QE28 A0A224Z4M0 A0A182HB54 A0A1W7RAP6 A0A0K8WKN7 A0A0K8UY29 A0A1A9UIX8 A0A1B0FMY9 E0W4C5 A0A1B6CY97 L7MII0 A0A1B6DAG0 D3TPY1 A0A026WLG8 A0A2M4BH05 A0A195CLC7 A0A2M4BHQ8 A0A182FIA1 A0A182J203 A0A1B0A9B8 A0A2M4BH22 A0A182VBE3 A0A1B0AKJ8 A0A0A1WGB5 A0A182TTD8 A0A1L8E9L3 A0A151XHZ1
A0A2A4JMM6 A0A194Q431 A0A212FIQ3 A0A0L7LTH0 A0A0M3QVG2 B4LKS1 A0A0M8ZNE6 A0A2A3E236 B4KQV8 K7JAD4 B4J9J3 A0A0C9QA03 B4P536 A0A1W4VDV6 A0A0K8TU52 A0A1L8E194 A0A1L8E1D4 A0A088A702 A0A0B4LFR4 A0A131XNK8 Q7K110 B4QBQ8 B4HN77 A0A232FBP6 A0A0L7RIL1 B3NLK0 B3MIC3 A0A067QE65 A0A2J7QDE8 A0A182NHK5 A0A154NZJ3 B0VZR0 A0A1L8EBK6 A0A1L8EC07 A0A1L8EBM7 A0A3B0J1P4 A0A1B0CEI4 A0A1I8NVP8 A0A1L8EBN3 T1PCA4 A0A1I8NG54 A0A1I8NG52 A0A023FDH8 A0A0J7LBH9 B5E0Y7 A0A1L8EBK7 B4GG23 A0A023GLS6 G3MM45 A0A023GNM8 A0A182MWC5 E2BN86 A0A023FWW6 A0A131XHF6 A0A0L0CEA5 A0A182I3Q6 B4NNG6 A0A182WIS6 A0A084VFB9 A0A182JNY7 A0A034WFW4 A0A182X6F3 A0A131Z110 A0A182P3I7 Q7QJS7 U5EVZ5 A0A182KRU4 A0A182QE28 A0A224Z4M0 A0A182HB54 A0A1W7RAP6 A0A0K8WKN7 A0A0K8UY29 A0A1A9UIX8 A0A1B0FMY9 E0W4C5 A0A1B6CY97 L7MII0 A0A1B6DAG0 D3TPY1 A0A026WLG8 A0A2M4BH05 A0A195CLC7 A0A2M4BHQ8 A0A182FIA1 A0A182J203 A0A1B0A9B8 A0A2M4BH22 A0A182VBE3 A0A1B0AKJ8 A0A0A1WGB5 A0A182TTD8 A0A1L8E9L3 A0A151XHZ1
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 18,
Likelihood: 0.985737
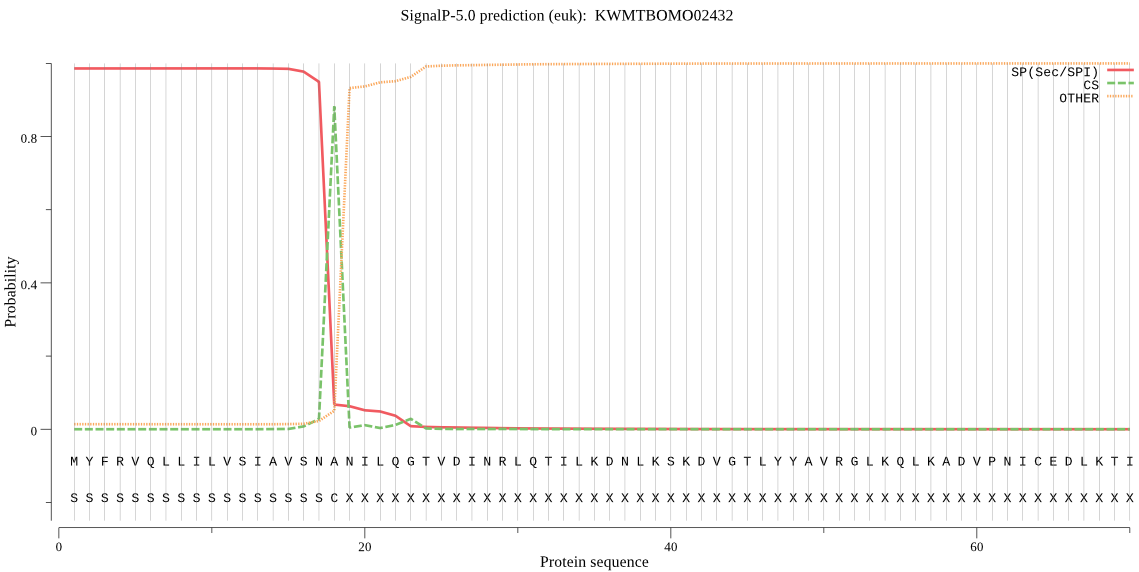
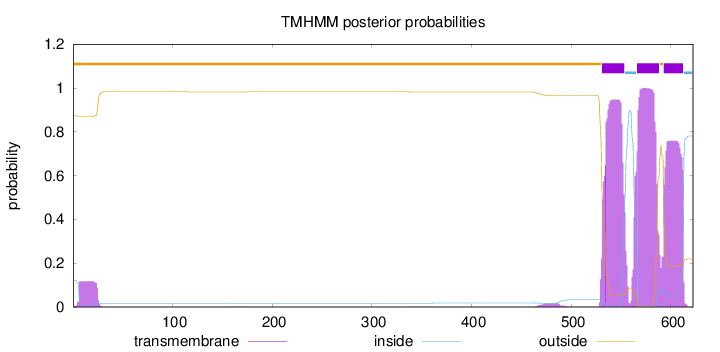
Length:
622
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
61.11959
Exp number, first 60 AAs:
2.30295
Total prob of N-in:
0.12538
outside
1 - 530
TMhelix
531 - 553
inside
554 - 565
TMhelix
566 - 588
outside
589 - 592
TMhelix
593 - 612
inside
613 - 622
Population Genetic Test Statistics
Pi
199.000183
Theta
175.860248
Tajima's D
-1.68396
CLR
1241.619157
CSRT
0.0417479126043698
Interpretation
Possibly Positive selection
