Pre Gene Modal
BGIBMGA013511
Annotation
H+_transporting_ATP_synthase_subunit_g_[Bombyx_mori]
Full name
ATP synthase subunit
Location in the cell
Mitochondrial Reliability : 1.849
Sequence
CDS
ATGGCCAGTGCAGTCGCTAAGGTGCCGACTCTTATTAATACTGCCATCACTCAGGCGAGGCCAAAATTAAATATTTTTATGAAATATGCTCGCGTTGAACTTGCTCCACCCAAGCTAAGTGAGCTGCCTCAGATTAGACAAGGCATCGGTAATCTGATCACAAGTGCTAAGACAGGAGCATGGAAAAGGCAAACAGTCAAAGAGGCCACACTAAATGTTCTGGTTGGAGCTGAAGTCATCTTCTGGTTCTACATTGGTGAATGCATTGGCAAACGTCACCTTGTTGGATACGATGTTTAA
Protein
MASAVAKVPTLINTAITQARPKLNIFMKYARVELAPPKLSELPQIRQGIGNLITSAKTGAWKRQTVKEATLNVLVGAEVIFWFYIGECIGKRHLVGYDV
Summary
Description
Mitochondrial membrane ATP synthase (F1F0 ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F1 - containing the extramembraneous catalytic core, and F0 - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F1 is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Part of the complex F0 domain. Minor subunit located with subunit a in the membrane.
Similarity
Belongs to the ATPase g subunit family.
Uniprot
Q1HQ25
A0A2H1W5F6
A0A2W1B8V1
A0A0K8SAR2
A0A2A4JL11
I4DMG4
+ More
E7EC47 I4DJX9 A0A0N1PHL7 A0A0L7KR56 A0A212FIR8 S4P8H9 A0A194Q436 T1D5J8 A0A1Z3GD37 A0A1S4J7A6 W8C2D6 B0W5Q6 A0A023ECN4 A0A1L8E1H1 A0A1L8EI22 A0A1L8E102 A0A1W4W424 B4HWY7 B4Q9W8 B3MJZ2 Q9VKM3 Q6XHY8 A0A182QD21 A0A182NEH3 A0A182JSZ6 Q1HR38 B3N504 B4LV35 A0A182IX31 A0A182Y5W3 B4JBE0 A0A182UXJ1 A0A182X0F1 Q7QG44 A0A182IE65 A0A0K8TQ25 A0A182PA82 V5G6W2 A0A0K8UY01 A0A1I8MJ31 A0A182VUX2 A0A182RGD2 A0A0A1WMN2 A0A034WLK6 A0A0C9QCF2 A0A1J1HEE5 A0A1I8P0H3 B4KHB6 A0A0L0BX87 B4G6Y6 Q29CN3 A0A336K1C2 A0A0M5J945 U5EU53 A0A1L8EF48 T1DF10 A0A2M4A6V0 A0A2M4C4L6 W5JCR9 A0A2M4A948 B4MU96 A0A2M3Z098 A0A182FWX8 A0A084VPJ2 D1FQD2 A0A1Y1KIS2 Q4PLY0 A0A2H8TN86 A0A224YMR4 A0A0C9QUF5 A0A131Z8H0 A0A131YBJ3 A0A0K8RN66 A0A023FGC0 T1GC57 A0A023FG13 L7M5S8 Q201Y2 A0A023GEC2 G3MSD9 A0A0K8RR45 A0A023FVZ4 V5HCJ3 A0A023FDR0 B7PSD2 A0A1W4XAY8 E2A8V7 C9W1F7 A0A1B0DB52 A0A2I9LNT0 A0A0C9QYT0 R4UX05 A0A2M4C4K0 A0A1E1WYK9
E7EC47 I4DJX9 A0A0N1PHL7 A0A0L7KR56 A0A212FIR8 S4P8H9 A0A194Q436 T1D5J8 A0A1Z3GD37 A0A1S4J7A6 W8C2D6 B0W5Q6 A0A023ECN4 A0A1L8E1H1 A0A1L8EI22 A0A1L8E102 A0A1W4W424 B4HWY7 B4Q9W8 B3MJZ2 Q9VKM3 Q6XHY8 A0A182QD21 A0A182NEH3 A0A182JSZ6 Q1HR38 B3N504 B4LV35 A0A182IX31 A0A182Y5W3 B4JBE0 A0A182UXJ1 A0A182X0F1 Q7QG44 A0A182IE65 A0A0K8TQ25 A0A182PA82 V5G6W2 A0A0K8UY01 A0A1I8MJ31 A0A182VUX2 A0A182RGD2 A0A0A1WMN2 A0A034WLK6 A0A0C9QCF2 A0A1J1HEE5 A0A1I8P0H3 B4KHB6 A0A0L0BX87 B4G6Y6 Q29CN3 A0A336K1C2 A0A0M5J945 U5EU53 A0A1L8EF48 T1DF10 A0A2M4A6V0 A0A2M4C4L6 W5JCR9 A0A2M4A948 B4MU96 A0A2M3Z098 A0A182FWX8 A0A084VPJ2 D1FQD2 A0A1Y1KIS2 Q4PLY0 A0A2H8TN86 A0A224YMR4 A0A0C9QUF5 A0A131Z8H0 A0A131YBJ3 A0A0K8RN66 A0A023FGC0 T1GC57 A0A023FG13 L7M5S8 Q201Y2 A0A023GEC2 G3MSD9 A0A0K8RR45 A0A023FVZ4 V5HCJ3 A0A023FDR0 B7PSD2 A0A1W4XAY8 E2A8V7 C9W1F7 A0A1B0DB52 A0A2I9LNT0 A0A0C9QYT0 R4UX05 A0A2M4C4K0 A0A1E1WYK9
Pubmed
19121390
28756777
22651552
26354079
26227816
22118469
+ More
23622113 24330624 24495485 24945155 17994087 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14525923 17550304 17204158 17510324 25244985 12364791 14747013 17210077 26369729 25315136 25830018 25348373 26108605 15632085 23185243 20920257 23761445 24438588 28004739 16431279 28797301 26830274 25576852 22216098 25765539 20798317 20650005 24029695 29248469 26131772 28503490
23622113 24330624 24495485 24945155 17994087 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14525923 17550304 17204158 17510324 25244985 12364791 14747013 17210077 26369729 25315136 25830018 25348373 26108605 15632085 23185243 20920257 23761445 24438588 28004739 16431279 28797301 26830274 25576852 22216098 25765539 20798317 20650005 24029695 29248469 26131772 28503490
EMBL
BABH01036046
DQ443227
ABF51316.1
ODYU01006430
SOQ48267.1
KZ150213
+ More
PZC72159.1 GBRD01015460 JAG50366.1 NWSH01001095 PCG72651.1 AK402482 BAM19104.1 HQ649757 ADV36663.1 AK401597 BAM18219.1 KQ460796 KPJ12176.1 JTDY01006695 KOB65767.1 AGBW02008342 OWR53619.1 GAIX01005921 JAA86639.1 KQ459465 KPJ00303.1 GALA01000473 JAA94379.1 KY783435 ASC55683.1 GAMC01000523 JAC06033.1 DS231844 EDS35709.1 GAPW01006480 JAC07118.1 GFDF01001739 JAV12345.1 GFDG01000540 JAV18259.1 GFDF01001740 JAV12344.1 CH480818 EDW52532.1 CM000361 CM002910 EDX04635.1 KMY89657.1 CH902620 EDV32447.1 KPU73942.1 AE014134 AY070693 AY071165 AAF53041.1 AAL48164.1 ABI31307.1 AY232044 CM000157 AAR10067.1 EDW88022.1 KRJ97523.1 AXCN02000858 DQ440256 CH477394 ABF18289.1 EAT41886.1 CH954177 EDV58949.1 CH940649 EDW63284.1 CH916368 EDW02945.1 AAAB01008839 EAA05846.3 APCN01004570 GDAI01001365 JAI16238.1 GALX01002641 JAB65825.1 GDHF01020767 JAI31547.1 GBXI01014170 JAD00122.1 GAKP01003408 JAC55544.1 GBYB01012123 JAG81890.1 CVRI01000001 CRK86327.1 CH933807 EDW13333.1 JRES01001205 KNC24620.1 CH479180 EDW28305.1 CH475442 EAL29399.1 UFQS01000055 UFQT01000055 UFQT01000338 SSW98716.1 SSX19102.1 SSX23340.1 CP012523 ALC39294.1 GANO01002478 JAB57393.1 GFDG01001481 JAV17318.1 GAMD01003296 JAA98294.1 GGFK01003164 MBW36485.1 GGFJ01011058 MBW60199.1 ADMH02001881 GGFL01004445 ETN60675.1 MBW68623.1 GGFK01003960 MBW37281.1 CH963857 EDW76022.1 GGFM01001196 MBW21947.1 ATLV01015007 KE524999 KFB39886.1 EZ420032 ACZ28387.1 GEZM01082553 JAV61323.1 DQ066349 AAY66986.1 GFXV01002933 MBW14738.1 GFPF01007761 MAA18907.1 GBYB01004262 JAG74029.1 GEDV01001309 JAP87248.1 GEFM01000340 JAP75456.1 GADI01001293 JAA72515.1 GBBK01004443 GBBK01004440 JAC20039.1 CAQQ02394156 GBBK01004442 JAC20040.1 GACK01006575 JAA58459.1 ABLF02028201 DQ413204 AK339998 ABD72672.1 BAH70677.1 GBBM01003191 JAC32227.1 JO844790 AEO36407.1 GADI01000415 JAA73393.1 GBBL01002397 JAC24923.1 GANP01009379 JAB75089.1 GBBK01004441 JAC20041.1 ABJB010110236 ABJB010266066 DS778027 EEC09504.1 GL437663 EFN70186.1 EZ406105 ACX53904.1 AJVK01000751 AJVK01000752 GFWZ01000052 MBW20042.1 GBZX01002815 JAG89925.1 KC741017 AGM32841.1 GGFJ01011126 MBW60267.1 GFAC01007079 JAT92109.1
PZC72159.1 GBRD01015460 JAG50366.1 NWSH01001095 PCG72651.1 AK402482 BAM19104.1 HQ649757 ADV36663.1 AK401597 BAM18219.1 KQ460796 KPJ12176.1 JTDY01006695 KOB65767.1 AGBW02008342 OWR53619.1 GAIX01005921 JAA86639.1 KQ459465 KPJ00303.1 GALA01000473 JAA94379.1 KY783435 ASC55683.1 GAMC01000523 JAC06033.1 DS231844 EDS35709.1 GAPW01006480 JAC07118.1 GFDF01001739 JAV12345.1 GFDG01000540 JAV18259.1 GFDF01001740 JAV12344.1 CH480818 EDW52532.1 CM000361 CM002910 EDX04635.1 KMY89657.1 CH902620 EDV32447.1 KPU73942.1 AE014134 AY070693 AY071165 AAF53041.1 AAL48164.1 ABI31307.1 AY232044 CM000157 AAR10067.1 EDW88022.1 KRJ97523.1 AXCN02000858 DQ440256 CH477394 ABF18289.1 EAT41886.1 CH954177 EDV58949.1 CH940649 EDW63284.1 CH916368 EDW02945.1 AAAB01008839 EAA05846.3 APCN01004570 GDAI01001365 JAI16238.1 GALX01002641 JAB65825.1 GDHF01020767 JAI31547.1 GBXI01014170 JAD00122.1 GAKP01003408 JAC55544.1 GBYB01012123 JAG81890.1 CVRI01000001 CRK86327.1 CH933807 EDW13333.1 JRES01001205 KNC24620.1 CH479180 EDW28305.1 CH475442 EAL29399.1 UFQS01000055 UFQT01000055 UFQT01000338 SSW98716.1 SSX19102.1 SSX23340.1 CP012523 ALC39294.1 GANO01002478 JAB57393.1 GFDG01001481 JAV17318.1 GAMD01003296 JAA98294.1 GGFK01003164 MBW36485.1 GGFJ01011058 MBW60199.1 ADMH02001881 GGFL01004445 ETN60675.1 MBW68623.1 GGFK01003960 MBW37281.1 CH963857 EDW76022.1 GGFM01001196 MBW21947.1 ATLV01015007 KE524999 KFB39886.1 EZ420032 ACZ28387.1 GEZM01082553 JAV61323.1 DQ066349 AAY66986.1 GFXV01002933 MBW14738.1 GFPF01007761 MAA18907.1 GBYB01004262 JAG74029.1 GEDV01001309 JAP87248.1 GEFM01000340 JAP75456.1 GADI01001293 JAA72515.1 GBBK01004443 GBBK01004440 JAC20039.1 CAQQ02394156 GBBK01004442 JAC20040.1 GACK01006575 JAA58459.1 ABLF02028201 DQ413204 AK339998 ABD72672.1 BAH70677.1 GBBM01003191 JAC32227.1 JO844790 AEO36407.1 GADI01000415 JAA73393.1 GBBL01002397 JAC24923.1 GANP01009379 JAB75089.1 GBBK01004441 JAC20041.1 ABJB010110236 ABJB010266066 DS778027 EEC09504.1 GL437663 EFN70186.1 EZ406105 ACX53904.1 AJVK01000751 AJVK01000752 GFWZ01000052 MBW20042.1 GBZX01002815 JAG89925.1 KC741017 AGM32841.1 GGFJ01011126 MBW60267.1 GFAC01007079 JAT92109.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000007151
UP000053268
+ More
UP000002320 UP000192221 UP000001292 UP000000304 UP000007801 UP000000803 UP000002282 UP000075886 UP000075884 UP000075881 UP000008820 UP000008711 UP000008792 UP000075880 UP000076408 UP000001070 UP000075903 UP000076407 UP000007062 UP000075840 UP000075885 UP000095301 UP000075920 UP000075900 UP000183832 UP000095300 UP000009192 UP000037069 UP000008744 UP000001819 UP000092553 UP000000673 UP000007798 UP000069272 UP000030765 UP000015102 UP000007819 UP000001555 UP000192223 UP000000311 UP000092462
UP000002320 UP000192221 UP000001292 UP000000304 UP000007801 UP000000803 UP000002282 UP000075886 UP000075884 UP000075881 UP000008820 UP000008711 UP000008792 UP000075880 UP000076408 UP000001070 UP000075903 UP000076407 UP000007062 UP000075840 UP000075885 UP000095301 UP000075920 UP000075900 UP000183832 UP000095300 UP000009192 UP000037069 UP000008744 UP000001819 UP000092553 UP000000673 UP000007798 UP000069272 UP000030765 UP000015102 UP000007819 UP000001555 UP000192223 UP000000311 UP000092462
Interpro
SUPFAM
SSF51182
SSF51182
ProteinModelPortal
Q1HQ25
A0A2H1W5F6
A0A2W1B8V1
A0A0K8SAR2
A0A2A4JL11
I4DMG4
+ More
E7EC47 I4DJX9 A0A0N1PHL7 A0A0L7KR56 A0A212FIR8 S4P8H9 A0A194Q436 T1D5J8 A0A1Z3GD37 A0A1S4J7A6 W8C2D6 B0W5Q6 A0A023ECN4 A0A1L8E1H1 A0A1L8EI22 A0A1L8E102 A0A1W4W424 B4HWY7 B4Q9W8 B3MJZ2 Q9VKM3 Q6XHY8 A0A182QD21 A0A182NEH3 A0A182JSZ6 Q1HR38 B3N504 B4LV35 A0A182IX31 A0A182Y5W3 B4JBE0 A0A182UXJ1 A0A182X0F1 Q7QG44 A0A182IE65 A0A0K8TQ25 A0A182PA82 V5G6W2 A0A0K8UY01 A0A1I8MJ31 A0A182VUX2 A0A182RGD2 A0A0A1WMN2 A0A034WLK6 A0A0C9QCF2 A0A1J1HEE5 A0A1I8P0H3 B4KHB6 A0A0L0BX87 B4G6Y6 Q29CN3 A0A336K1C2 A0A0M5J945 U5EU53 A0A1L8EF48 T1DF10 A0A2M4A6V0 A0A2M4C4L6 W5JCR9 A0A2M4A948 B4MU96 A0A2M3Z098 A0A182FWX8 A0A084VPJ2 D1FQD2 A0A1Y1KIS2 Q4PLY0 A0A2H8TN86 A0A224YMR4 A0A0C9QUF5 A0A131Z8H0 A0A131YBJ3 A0A0K8RN66 A0A023FGC0 T1GC57 A0A023FG13 L7M5S8 Q201Y2 A0A023GEC2 G3MSD9 A0A0K8RR45 A0A023FVZ4 V5HCJ3 A0A023FDR0 B7PSD2 A0A1W4XAY8 E2A8V7 C9W1F7 A0A1B0DB52 A0A2I9LNT0 A0A0C9QYT0 R4UX05 A0A2M4C4K0 A0A1E1WYK9
E7EC47 I4DJX9 A0A0N1PHL7 A0A0L7KR56 A0A212FIR8 S4P8H9 A0A194Q436 T1D5J8 A0A1Z3GD37 A0A1S4J7A6 W8C2D6 B0W5Q6 A0A023ECN4 A0A1L8E1H1 A0A1L8EI22 A0A1L8E102 A0A1W4W424 B4HWY7 B4Q9W8 B3MJZ2 Q9VKM3 Q6XHY8 A0A182QD21 A0A182NEH3 A0A182JSZ6 Q1HR38 B3N504 B4LV35 A0A182IX31 A0A182Y5W3 B4JBE0 A0A182UXJ1 A0A182X0F1 Q7QG44 A0A182IE65 A0A0K8TQ25 A0A182PA82 V5G6W2 A0A0K8UY01 A0A1I8MJ31 A0A182VUX2 A0A182RGD2 A0A0A1WMN2 A0A034WLK6 A0A0C9QCF2 A0A1J1HEE5 A0A1I8P0H3 B4KHB6 A0A0L0BX87 B4G6Y6 Q29CN3 A0A336K1C2 A0A0M5J945 U5EU53 A0A1L8EF48 T1DF10 A0A2M4A6V0 A0A2M4C4L6 W5JCR9 A0A2M4A948 B4MU96 A0A2M3Z098 A0A182FWX8 A0A084VPJ2 D1FQD2 A0A1Y1KIS2 Q4PLY0 A0A2H8TN86 A0A224YMR4 A0A0C9QUF5 A0A131Z8H0 A0A131YBJ3 A0A0K8RN66 A0A023FGC0 T1GC57 A0A023FG13 L7M5S8 Q201Y2 A0A023GEC2 G3MSD9 A0A0K8RR45 A0A023FVZ4 V5HCJ3 A0A023FDR0 B7PSD2 A0A1W4XAY8 E2A8V7 C9W1F7 A0A1B0DB52 A0A2I9LNT0 A0A0C9QYT0 R4UX05 A0A2M4C4K0 A0A1E1WYK9
Ontologies
PATHWAY
GO
PANTHER
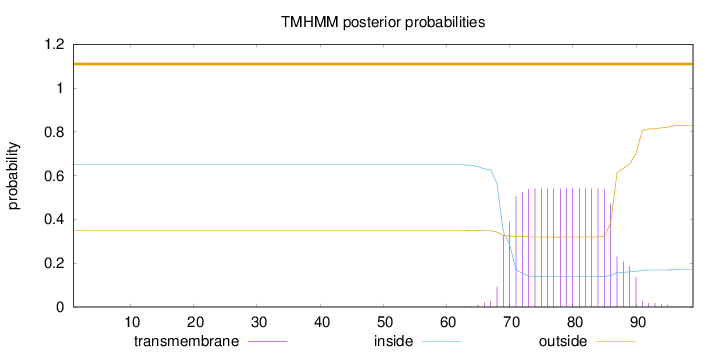
Topology
Subcellular location
Mitochondrion
Length:
99
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.25559
Exp number, first 60 AAs:
0.00599999999999999
Total prob of N-in:
0.64952
outside
1 - 99
Population Genetic Test Statistics
Pi
263.678236
Theta
192.569295
Tajima's D
1.162143
CLR
0
CSRT
0.703114844257787
Interpretation
Uncertain
