Pre Gene Modal
BGIBMGA013512
Annotation
PREDICTED:_neuroblastoma-amplified_sequence-like_[Bombyx_mori]
Full name
Neuroblastoma-amplified sequence
Alternative Name
Neuroblastoma-amplified gene protein homolog
Location in the cell
Cytoplasmic Reliability : 3.37
Sequence
CDS
ATGGCCGCCGAGCAATGTGACCAAAAGGAGGCATTGGCGAGTGAAGAAGGTCGACGTGAATTATTCGAGAAGTTCCTTGAGGAGTCCGACACTTGGCAACAGAGGAAGGCTCTGGTCGACGTGCTGAACTGCTGGCCTCCGATTACTTCCAGCGAGTCGGCGAGTCTGCAGTGCGAGTACCTGTACGCGCTGCTGACGAACCCTCTGGACAGCAAGGAGAGCTTGGAGCTCATCAAGGCGCTGCTCAGCCGGCCCGTCCTTAGCGAGCAGGAAATTAAATGGCTGATCGACGACGTCGTCCCGGACGCAGTCGTGAACGCCATCTGGATTCTCTTGCTCAGTAAATATCAACATTCCAAAGAAGAGATACTGAACATATTGCAACAGCACAAGGAAGAAGTTGCGAAACAAGAAATCGACGATGAGCTGGTCGCGGAGTTGCTCCGCAACGACTTGTTCGTGAGTCTGGTCGGCACTCCGCTGTACCCCAAAATGCTCAGCTACATCATCAGCCGGGAGGGCGGCTCCGAGGACGCGCCGTACTCCGTGCCGTGGGCCACGCGAGAGCTGCTGAAGGCCAACTACCTGGCCGAGGCCGGGCACCTCCGTCTCATGGCTGATGGCGTCCCGCCGCCCCTGCGAGGGTTCTCTCAGTCCGTTCTCTATTGTAAGAATATCTTTAAGAAATAA
Protein
MAAEQCDQKEALASEEGRRELFEKFLEESDTWQQRKALVDVLNCWPPITSSESASLQCEYLYALLTNPLDSKESLELIKALLSRPVLSEQEIKWLIDDVVPDAVVNAIWILLLSKYQHSKEEILNILQQHKEEVAKQEIDDELVAELLRNDLFVSLVGTPLYPKMLSYIISREGGSEDAPYSVPWATRELLKANYLAEAGHLRLMADGVPPPLRGFSQSVLYCKNIFKK
Summary
Description
Involved in Golgi-to-endoplasmic reticulum (ER) retrograde transport; the function is proposed to depend on its association in the NRZ complex which is believed to play a role in SNARE assembly at the ER (By similarity).
Subunit
Component of the NRZ complex.
Keywords
Coiled coil
Complete proteome
Endoplasmic reticulum
Protein transport
Reference proteome
Repeat
Transport
WD repeat
Feature
chain Neuroblastoma-amplified sequence
Uniprot
H9JVE9
A0A2A4JMH6
A0A2W1BDD6
A0A1E1WTL1
A0A194Q4M9
A0A212FIR2
+ More
S4P6G8 A0A0L8GAE7 A0A0L8GA97 A0A210Q6V8 A0A162QEA8 A0A0N8AHJ0 A0A0P5UQI9 A0A0P5XMP1 A0A0P5BBN8 A0A0P6JA84 A0A0P5RID4 A0A0P5T3B6 A0A0N8CIV4 A0A0N8DQ35 A0A0P5FBS2 A0A0P5N4N4 A0A0P5PE49 A0A0P6E266 A0A0P5Q9F4 F7AJ11 V4AB70 R7T8S2 E9H0R3 A0A0P5YGX3 A0A0P5YE61 Q5TYW4 A0A0P5A7L8 A0A1B6DSA4 W5UEB5 A0A2D0T1M5 C3Y2C0 A0A3B3SPE2 A0A2I0UL20 W5NI89 K7GA65 K7GA49 A0A2J8V472 F7G1Z4 F7G1Z9 A0A131YVD2 A0A1D5Q3E8 A0A1L8GBP3 I3IVK2 K9IPL1 A0A2C9KR62 H7BZR3 A0A093P9J3 A0A0P5Y316 A0A3B3DQK4 A0A091JAM3 A0A1D5PK79 H3DFW0 A0A3B4DJ64 A0A1D5P365 A0A3P8P8Z5 A0A3P8P8K8 A0A3P8P880 A0A3P9LQ30 A0A3P8P9A2 Q80XK8 A0A3P9CDM3 A0A093HCD6 Q6PGH6 Q6GQV6 G1NML7 Q3UJX6 E9Q411 Q80X24 A0A0A0AJB7 H2MQ52 A0A3Q0G330 A0A1S3JM80 A0A091MRC3 A0A3B3Z8K3 A0A3P8ZLY7 A0A3B4BNR5 A0A1S3KGU9 G3W2R0 H3A3U3 A0A091V4N4 A0A091WDX1 A0A2L2YH20 A0A2J8NLR1
S4P6G8 A0A0L8GAE7 A0A0L8GA97 A0A210Q6V8 A0A162QEA8 A0A0N8AHJ0 A0A0P5UQI9 A0A0P5XMP1 A0A0P5BBN8 A0A0P6JA84 A0A0P5RID4 A0A0P5T3B6 A0A0N8CIV4 A0A0N8DQ35 A0A0P5FBS2 A0A0P5N4N4 A0A0P5PE49 A0A0P6E266 A0A0P5Q9F4 F7AJ11 V4AB70 R7T8S2 E9H0R3 A0A0P5YGX3 A0A0P5YE61 Q5TYW4 A0A0P5A7L8 A0A1B6DSA4 W5UEB5 A0A2D0T1M5 C3Y2C0 A0A3B3SPE2 A0A2I0UL20 W5NI89 K7GA65 K7GA49 A0A2J8V472 F7G1Z4 F7G1Z9 A0A131YVD2 A0A1D5Q3E8 A0A1L8GBP3 I3IVK2 K9IPL1 A0A2C9KR62 H7BZR3 A0A093P9J3 A0A0P5Y316 A0A3B3DQK4 A0A091JAM3 A0A1D5PK79 H3DFW0 A0A3B4DJ64 A0A1D5P365 A0A3P8P8Z5 A0A3P8P8K8 A0A3P8P880 A0A3P9LQ30 A0A3P8P9A2 Q80XK8 A0A3P9CDM3 A0A093HCD6 Q6PGH6 Q6GQV6 G1NML7 Q3UJX6 E9Q411 Q80X24 A0A0A0AJB7 H2MQ52 A0A3Q0G330 A0A1S3JM80 A0A091MRC3 A0A3B3Z8K3 A0A3P8ZLY7 A0A3B4BNR5 A0A1S3KGU9 G3W2R0 H3A3U3 A0A091V4N4 A0A091WDX1 A0A2L2YH20 A0A2J8NLR1
Pubmed
19121390
28756777
26354079
22118469
23622113
28812685
+ More
20431018 23254933 21292972 23594743 23127152 18563158 29240929 17381049 18464734 26830274 17431167 27762356 25186727 15562597 15815621 29451363 15592404 15496914 17554307 15489334 20838655 10349636 11042159 11076861 11217851 12466851 16141073 17242355 19468303 21183079 25463417 25069045 26383154 21709235 9215903 26561354
20431018 23254933 21292972 23594743 23127152 18563158 29240929 17381049 18464734 26830274 17431167 27762356 25186727 15562597 15815621 29451363 15592404 15496914 17554307 15489334 20838655 10349636 11042159 11076861 11217851 12466851 16141073 17242355 19468303 21183079 25463417 25069045 26383154 21709235 9215903 26561354
EMBL
BABH01036050
BABH01036051
BABH01036052
BABH01036053
BABH01036054
BABH01036055
+ More
NWSH01001095 PCG72633.1 KZ150118 PZC73232.1 GDQN01000777 JAT90277.1 KQ459465 KPJ00304.1 AGBW02008342 OWR53625.1 GAIX01005044 JAA87516.1 KQ422986 KOF73824.1 KOF73823.1 NEDP02004772 OWF44470.1 LRGB01000341 KZS19617.1 GDIP01146817 JAJ76585.1 GDIP01113317 JAL90397.1 GDIP01069875 JAM33840.1 GDIP01186712 JAJ36690.1 GDIQ01020565 JAN74172.1 GDIQ01101166 JAL50560.1 GDIP01133965 JAL69749.1 GDIP01127415 JAL76299.1 GDIP01011967 JAM91748.1 GDIP01149518 JAJ73884.1 GDIQ01147101 JAL04625.1 GDIQ01132065 JAL19661.1 GDIQ01068880 JAN25857.1 GDIQ01117709 JAL34017.1 AAMC01114030 AAMC01114031 AAMC01114032 AAMC01114033 AAMC01114034 AAMC01114035 AAMC01114036 AAMC01114037 AAMC01114038 AAMC01114039 KB200521 ESP01249.1 AMQN01014563 AMQN01014564 KB311057 ELT90143.1 GL732581 EFX74704.1 GDIP01058133 JAM45582.1 GDIP01060426 JAM43289.1 BX649472 BX649264 BX649299 BC135054 GDIP01203007 JAJ20395.1 GEDC01008779 JAS28519.1 JT412617 AHH39924.1 GG666480 EEN66010.1 KZ505701 PKU46708.1 AHAT01009097 AHAT01009098 AHAT01009099 AHAT01009100 AHAT01009101 AHAT01009102 AHAT01009103 AGCU01198987 AGCU01198988 AGCU01198989 AGCU01198990 AGCU01198991 AGCU01198992 AGCU01198993 AGCU01198994 AGCU01198995 AGCU01198996 NDHI03003434 PNJ52334.1 AAPN01436104 AAPN01436105 AAPN01436106 AAPN01436107 AAPN01436108 AAPN01436109 AAPN01436110 AAPN01436111 AAPN01436112 AAPN01436113 GEDV01006117 JAP82440.1 JSUE03010259 JSUE03010260 JSUE03010261 JSUE03010262 CM004474 OCT81253.1 AERX01005424 AERX01005425 AERX01005426 AERX01005427 AERX01005428 GABZ01003407 JAA50118.1 AC007738 AC008278 AC008282 AC074184 KL225502 KFW73071.1 GDIP01064628 JAM39087.1 KK501620 KFP16770.1 AADN05000843 BC046406 AAH46406.1 KL217400 KFV77090.1 BC057020 AAH57020.1 BC072598 AAH72598.1 AK146270 BAE27029.1 AC154517 AC159250 CT025539 CT030206 BC051652 AAH51652.1 KL871774 KGL93548.1 KK833705 KFP78461.1 AEFK01036733 AEFK01036734 AEFK01036735 AEFK01036736 AEFK01036737 AEFK01036738 AEFK01036739 AEFK01036740 AEFK01036741 AEFK01036742 AFYH01168741 AFYH01168742 AFYH01168743 AFYH01168744 AFYH01168745 AFYH01168746 AFYH01168747 AFYH01168748 AFYH01168749 AFYH01168750 KL410622 KFQ97953.1 KK735179 KFR12983.1 IAAA01029226 LAA07456.1 NBAG03000227 PNI72710.1
NWSH01001095 PCG72633.1 KZ150118 PZC73232.1 GDQN01000777 JAT90277.1 KQ459465 KPJ00304.1 AGBW02008342 OWR53625.1 GAIX01005044 JAA87516.1 KQ422986 KOF73824.1 KOF73823.1 NEDP02004772 OWF44470.1 LRGB01000341 KZS19617.1 GDIP01146817 JAJ76585.1 GDIP01113317 JAL90397.1 GDIP01069875 JAM33840.1 GDIP01186712 JAJ36690.1 GDIQ01020565 JAN74172.1 GDIQ01101166 JAL50560.1 GDIP01133965 JAL69749.1 GDIP01127415 JAL76299.1 GDIP01011967 JAM91748.1 GDIP01149518 JAJ73884.1 GDIQ01147101 JAL04625.1 GDIQ01132065 JAL19661.1 GDIQ01068880 JAN25857.1 GDIQ01117709 JAL34017.1 AAMC01114030 AAMC01114031 AAMC01114032 AAMC01114033 AAMC01114034 AAMC01114035 AAMC01114036 AAMC01114037 AAMC01114038 AAMC01114039 KB200521 ESP01249.1 AMQN01014563 AMQN01014564 KB311057 ELT90143.1 GL732581 EFX74704.1 GDIP01058133 JAM45582.1 GDIP01060426 JAM43289.1 BX649472 BX649264 BX649299 BC135054 GDIP01203007 JAJ20395.1 GEDC01008779 JAS28519.1 JT412617 AHH39924.1 GG666480 EEN66010.1 KZ505701 PKU46708.1 AHAT01009097 AHAT01009098 AHAT01009099 AHAT01009100 AHAT01009101 AHAT01009102 AHAT01009103 AGCU01198987 AGCU01198988 AGCU01198989 AGCU01198990 AGCU01198991 AGCU01198992 AGCU01198993 AGCU01198994 AGCU01198995 AGCU01198996 NDHI03003434 PNJ52334.1 AAPN01436104 AAPN01436105 AAPN01436106 AAPN01436107 AAPN01436108 AAPN01436109 AAPN01436110 AAPN01436111 AAPN01436112 AAPN01436113 GEDV01006117 JAP82440.1 JSUE03010259 JSUE03010260 JSUE03010261 JSUE03010262 CM004474 OCT81253.1 AERX01005424 AERX01005425 AERX01005426 AERX01005427 AERX01005428 GABZ01003407 JAA50118.1 AC007738 AC008278 AC008282 AC074184 KL225502 KFW73071.1 GDIP01064628 JAM39087.1 KK501620 KFP16770.1 AADN05000843 BC046406 AAH46406.1 KL217400 KFV77090.1 BC057020 AAH57020.1 BC072598 AAH72598.1 AK146270 BAE27029.1 AC154517 AC159250 CT025539 CT030206 BC051652 AAH51652.1 KL871774 KGL93548.1 KK833705 KFP78461.1 AEFK01036733 AEFK01036734 AEFK01036735 AEFK01036736 AEFK01036737 AEFK01036738 AEFK01036739 AEFK01036740 AEFK01036741 AEFK01036742 AFYH01168741 AFYH01168742 AFYH01168743 AFYH01168744 AFYH01168745 AFYH01168746 AFYH01168747 AFYH01168748 AFYH01168749 AFYH01168750 KL410622 KFQ97953.1 KK735179 KFR12983.1 IAAA01029226 LAA07456.1 NBAG03000227 PNI72710.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053454
UP000242188
+ More
UP000076858 UP000008143 UP000030746 UP000014760 UP000000305 UP000000437 UP000221080 UP000001554 UP000261540 UP000018468 UP000007267 UP000002279 UP000006718 UP000186698 UP000005207 UP000076420 UP000005640 UP000054081 UP000261560 UP000053119 UP000000539 UP000007303 UP000261440 UP000265100 UP000265180 UP000265160 UP000053875 UP000001645 UP000000589 UP000053858 UP000001038 UP000189705 UP000085678 UP000261520 UP000265140 UP000007648 UP000008672 UP000053283 UP000053605
UP000076858 UP000008143 UP000030746 UP000014760 UP000000305 UP000000437 UP000221080 UP000001554 UP000261540 UP000018468 UP000007267 UP000002279 UP000006718 UP000186698 UP000005207 UP000076420 UP000005640 UP000054081 UP000261560 UP000053119 UP000000539 UP000007303 UP000261440 UP000265100 UP000265180 UP000265160 UP000053875 UP000001645 UP000000589 UP000053858 UP000001038 UP000189705 UP000085678 UP000261520 UP000265140 UP000007648 UP000008672 UP000053283 UP000053605
Interpro
Gene 3D
ProteinModelPortal
H9JVE9
A0A2A4JMH6
A0A2W1BDD6
A0A1E1WTL1
A0A194Q4M9
A0A212FIR2
+ More
S4P6G8 A0A0L8GAE7 A0A0L8GA97 A0A210Q6V8 A0A162QEA8 A0A0N8AHJ0 A0A0P5UQI9 A0A0P5XMP1 A0A0P5BBN8 A0A0P6JA84 A0A0P5RID4 A0A0P5T3B6 A0A0N8CIV4 A0A0N8DQ35 A0A0P5FBS2 A0A0P5N4N4 A0A0P5PE49 A0A0P6E266 A0A0P5Q9F4 F7AJ11 V4AB70 R7T8S2 E9H0R3 A0A0P5YGX3 A0A0P5YE61 Q5TYW4 A0A0P5A7L8 A0A1B6DSA4 W5UEB5 A0A2D0T1M5 C3Y2C0 A0A3B3SPE2 A0A2I0UL20 W5NI89 K7GA65 K7GA49 A0A2J8V472 F7G1Z4 F7G1Z9 A0A131YVD2 A0A1D5Q3E8 A0A1L8GBP3 I3IVK2 K9IPL1 A0A2C9KR62 H7BZR3 A0A093P9J3 A0A0P5Y316 A0A3B3DQK4 A0A091JAM3 A0A1D5PK79 H3DFW0 A0A3B4DJ64 A0A1D5P365 A0A3P8P8Z5 A0A3P8P8K8 A0A3P8P880 A0A3P9LQ30 A0A3P8P9A2 Q80XK8 A0A3P9CDM3 A0A093HCD6 Q6PGH6 Q6GQV6 G1NML7 Q3UJX6 E9Q411 Q80X24 A0A0A0AJB7 H2MQ52 A0A3Q0G330 A0A1S3JM80 A0A091MRC3 A0A3B3Z8K3 A0A3P8ZLY7 A0A3B4BNR5 A0A1S3KGU9 G3W2R0 H3A3U3 A0A091V4N4 A0A091WDX1 A0A2L2YH20 A0A2J8NLR1
S4P6G8 A0A0L8GAE7 A0A0L8GA97 A0A210Q6V8 A0A162QEA8 A0A0N8AHJ0 A0A0P5UQI9 A0A0P5XMP1 A0A0P5BBN8 A0A0P6JA84 A0A0P5RID4 A0A0P5T3B6 A0A0N8CIV4 A0A0N8DQ35 A0A0P5FBS2 A0A0P5N4N4 A0A0P5PE49 A0A0P6E266 A0A0P5Q9F4 F7AJ11 V4AB70 R7T8S2 E9H0R3 A0A0P5YGX3 A0A0P5YE61 Q5TYW4 A0A0P5A7L8 A0A1B6DSA4 W5UEB5 A0A2D0T1M5 C3Y2C0 A0A3B3SPE2 A0A2I0UL20 W5NI89 K7GA65 K7GA49 A0A2J8V472 F7G1Z4 F7G1Z9 A0A131YVD2 A0A1D5Q3E8 A0A1L8GBP3 I3IVK2 K9IPL1 A0A2C9KR62 H7BZR3 A0A093P9J3 A0A0P5Y316 A0A3B3DQK4 A0A091JAM3 A0A1D5PK79 H3DFW0 A0A3B4DJ64 A0A1D5P365 A0A3P8P8Z5 A0A3P8P8K8 A0A3P8P880 A0A3P9LQ30 A0A3P8P9A2 Q80XK8 A0A3P9CDM3 A0A093HCD6 Q6PGH6 Q6GQV6 G1NML7 Q3UJX6 E9Q411 Q80X24 A0A0A0AJB7 H2MQ52 A0A3Q0G330 A0A1S3JM80 A0A091MRC3 A0A3B3Z8K3 A0A3P8ZLY7 A0A3B4BNR5 A0A1S3KGU9 G3W2R0 H3A3U3 A0A091V4N4 A0A091WDX1 A0A2L2YH20 A0A2J8NLR1
Ontologies
KEGG
GO
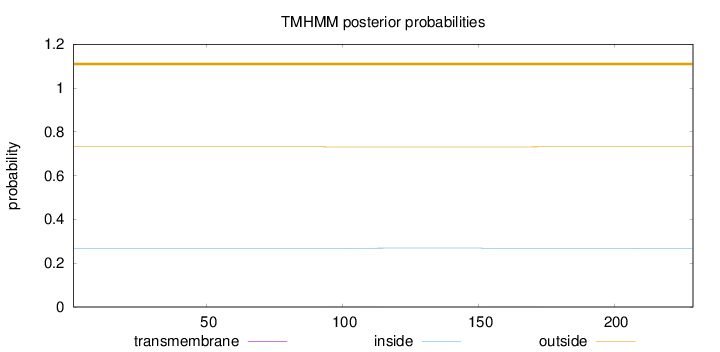
Topology
Subcellular location
Endoplasmic reticulum
Length:
229
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03253
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.26862
outside
1 - 229
Population Genetic Test Statistics
Pi
199.076333
Theta
164.704377
Tajima's D
0.917229
CLR
0.07914
CSRT
0.635418229088546
Interpretation
Uncertain
