Gene
KWMTBOMO02422 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013493
Annotation
phosphohistidine_phosphatase_[Bombyx_mori]
Full name
Sex-regulated protein janus-A
+ More
14 kDa phosphohistidine phosphatase
14 kDa phosphohistidine phosphatase
Alternative Name
Phosphohistidine phosphatase 1
Protein histidine phosphatase
Protein histidine phosphatase
Location in the cell
Cytoplasmic Reliability : 1.447 Mitochondrial Reliability : 1.454
Sequence
CDS
ATGCCTTTTAATCCTTTCAAATTATTTAGTCGACCCCGAGTAAATAATACCAAATTCTGGAACCCAAATTTATCAGGGCCAATATTACAATTTACCACAAAAATGTCTGGCTCACTCCACAAAGTTCCTAAAGTGGACATTGATAAATCTGGAGTATTCAAGTACATCCTCCTGAAGGTCTATGACCAAGAAGTTGATGGAGTGGAACCTTCCACTATTGTCTTAAGAGGATATAAGAGATGCAACTATCACTCTGATATATATGATGAGGTACAAGGTAAATTAATCCCCTTTGATTGTGAGCCTTGGGGTGGTGGTAGAATATCTCATGATCCAGACAATAAGAAGATACACATCTATGGATACTCTCAGGCCTATGGAAAGGCAGACCATGAGGAGGCAGCCAAACTAATCAAGAATGCATATCCAGATTACACAATCACAATCAGTGATGAAGGATACTAA
Protein
MPFNPFKLFSRPRVNNTKFWNPNLSGPILQFTTKMSGSLHKVPKVDIDKSGVFKYILLKVYDQEVDGVEPSTIVLRGYKRCNYHSDIYDEVQGKLIPFDCEPWGGGRISHDPDNKKIHIYGYSQAYGKADHEEAAKLIKNAYPDYTITISDEGY
Summary
Description
JanA and janB regulate somatic sex differentiation.
Exhibits phosphohistidine phosphatase activity.
Exhibits phosphohistidine phosphatase activity.
Catalytic Activity
H2O + N(pros)-phospho-L-histidyl-[protein] = L-histidyl-[protein] + phosphate
H2O + N(tele)-phospho-L-histidyl-[protein] = L-histidyl-[protein] + phosphate
H2O + N(tele)-phospho-L-histidyl-[protein] = L-histidyl-[protein] + phosphate
Subunit
Monomer.
Similarity
Belongs to the janus family.
Keywords
Complete proteome
Differentiation
Reference proteome
Sexual differentiation
Cytoplasm
Hydrolase
Protein phosphatase
Acetylation
Feature
chain Sex-regulated protein janus-A
Uniprot
Q2F5N5
H9JVD0
A0A1E1VX92
A0A2H1VDJ4
A0A2W1BK75
A0A2A4JLE7
+ More
S4PGJ4 A0A0L7L2G8 A0A212F5L1 A0A194Q4N4 A0A0N1PGL2 A0A0N1PI35 A0A182R4T0 A0A1I8NB48 A0A1B6LYP8 A0A2M4CZN2 A0A0V0GDK6 A0A182VWL1 A0A182YHG7 A0A182T9L2 A0A1B6H1Q8 A0A2M4AY21 A0A069DP21 A0A2M3ZDK6 A0A182FNZ3 A0A2M4C3L3 A0A336MDK8 A0A182MKI6 A0A088ACD4 A0A2M4AY66 A0A182HZZ2 A0A3B3U8L0 A0A182Q8C2 A0A182P575 A0A182N5U6 A0A182ILD5 A0A0K8THC0 A0A0A9ZD48 A0A0L0CHZ4 I5AMV9 P54364 A0A0P4VLC5 R4FMN8 A0A023F860 A0A182V6Q2 A0A182TJU1 A0A182LR35 A0A182X0A2 Q7PUI9 A0A224XWS2 A0A2M4CTP6 A0A1B6HP61 A0A1S4F7C0 A0A182JVI0 A0A3B3WX09 A0A087X7T5 A0A087X5G2 B4NJJ3 B4GNH7 D0QWR6 U5EVH9 A0A0S7LEG5 A0A146R1V4 A0A3B0K7G7 A0A147A8G3 M3ZLH2 A0A2A3EHH3 A0A182G4V8 G3IHB0 F6WVK9 A0A0M4F3P7 Q17D02 A0A1Q3FBA0 B0WP95 A0A1A9WT60 A0A226NGC1 A0A226PMI4 A0A2P4S9G9 H0XUK7 Q9DAK9 A0A1A9VDH0 A0A1D5PWG8 D3ZP47 A0A3B4ANQ6 A0A250Y9L6 A0A3B4AME5 A0A0A9X5P1 A0A023EDZ0 I3NEJ4 A0A2K6EIY3 G3Q237 A0A3P9K6C4 A0A3B3H9F4 A0A3B3C695 R7U055 Q32PA4 A0A0M3KB29 A0A1U7R1F2 A0A3P9JMV2 A0A0K8TQA2
S4PGJ4 A0A0L7L2G8 A0A212F5L1 A0A194Q4N4 A0A0N1PGL2 A0A0N1PI35 A0A182R4T0 A0A1I8NB48 A0A1B6LYP8 A0A2M4CZN2 A0A0V0GDK6 A0A182VWL1 A0A182YHG7 A0A182T9L2 A0A1B6H1Q8 A0A2M4AY21 A0A069DP21 A0A2M3ZDK6 A0A182FNZ3 A0A2M4C3L3 A0A336MDK8 A0A182MKI6 A0A088ACD4 A0A2M4AY66 A0A182HZZ2 A0A3B3U8L0 A0A182Q8C2 A0A182P575 A0A182N5U6 A0A182ILD5 A0A0K8THC0 A0A0A9ZD48 A0A0L0CHZ4 I5AMV9 P54364 A0A0P4VLC5 R4FMN8 A0A023F860 A0A182V6Q2 A0A182TJU1 A0A182LR35 A0A182X0A2 Q7PUI9 A0A224XWS2 A0A2M4CTP6 A0A1B6HP61 A0A1S4F7C0 A0A182JVI0 A0A3B3WX09 A0A087X7T5 A0A087X5G2 B4NJJ3 B4GNH7 D0QWR6 U5EVH9 A0A0S7LEG5 A0A146R1V4 A0A3B0K7G7 A0A147A8G3 M3ZLH2 A0A2A3EHH3 A0A182G4V8 G3IHB0 F6WVK9 A0A0M4F3P7 Q17D02 A0A1Q3FBA0 B0WP95 A0A1A9WT60 A0A226NGC1 A0A226PMI4 A0A2P4S9G9 H0XUK7 Q9DAK9 A0A1A9VDH0 A0A1D5PWG8 D3ZP47 A0A3B4ANQ6 A0A250Y9L6 A0A3B4AME5 A0A0A9X5P1 A0A023EDZ0 I3NEJ4 A0A2K6EIY3 G3Q237 A0A3P9K6C4 A0A3B3H9F4 A0A3B3C695 R7U055 Q32PA4 A0A0M3KB29 A0A1U7R1F2 A0A3P9JMV2 A0A0K8TQA2
EC Number
3.9.1.3
Pubmed
19121390
28756777
23622113
26227816
22118469
26354079
+ More
25315136 25244985 26334808 25401762 26823975 26108605 15632085 17994087 7700229 27129103 25474469 20966253 12364791 19859648 23542700 26483478 21804562 29704459 17495919 17510324 24621616 16141072 15489334 21183079 15592404 15057822 15632090 22673903 25463417 28087693 24945155 17554307 29451363 23254933 26369729
25315136 25244985 26334808 25401762 26823975 26108605 15632085 17994087 7700229 27129103 25474469 20966253 12364791 19859648 23542700 26483478 21804562 29704459 17495919 17510324 24621616 16141072 15489334 21183079 15592404 15057822 15632090 22673903 25463417 28087693 24945155 17554307 29451363 23254933 26369729
EMBL
DQ311388
ABD36332.1
BABH01036055
GDQN01011706
JAT79348.1
ODYU01001972
+ More
SOQ38909.1 KZ150118 PZC73236.1 NWSH01001095 PCG72639.1 GAIX01006190 JAA86370.1 JTDY01003397 KOB69620.1 AGBW02010179 OWR49018.1 KQ459465 KPJ00309.1 KQ460393 KPJ15379.1 LADJ01008100 KPJ20915.1 GEBQ01018260 GEBQ01011159 JAT21717.1 JAT28818.1 GGFL01006614 MBW70792.1 GECL01000722 JAP05402.1 GECZ01001160 JAS68609.1 GGFK01012324 MBW45645.1 GBGD01003493 JAC85396.1 GGFM01005820 MBW26571.1 GGFJ01010752 MBW59893.1 UFQT01000991 SSX28326.1 AXCM01002444 GGFK01012207 MBW45528.1 APCN01002026 AXCN02001153 GBRD01001291 JAG64530.1 GBHO01031187 GBHO01031186 GBHO01031185 GBHO01031183 GBHO01003881 GBHO01003879 GDHC01015863 GDHC01005617 JAG12417.1 JAG12418.1 JAG12419.1 JAG12421.1 JAG39723.1 JAG39725.1 JAQ02766.1 JAQ13012.1 JRES01000378 KNC31832.1 CM000070 EIM52294.1 S77099 GDKW01003164 JAI53431.1 GAHY01000908 JAA76602.1 GBBI01001275 JAC17437.1 AAAB01008987 EAA01434.5 GFTR01002098 JAW14328.1 GGFL01004544 MBW68722.1 GECU01031236 JAS76470.1 AYCK01016959 AYCK01026587 CH964272 EDW83917.1 CH479186 EDW39303.1 FJ821034 ACN94747.1 GANO01003398 JAB56473.1 GBYX01173258 JAO87841.1 GCES01123887 JAQ62435.1 OUUW01000005 SPP80941.1 GCES01011505 JAR74818.1 KZ288256 PBC30636.1 JXUM01144562 KQ569817 KXJ68510.1 JH002744 RAZU01000235 EGW14296.1 RLQ63888.1 CP012526 ALC46262.1 CH477301 EAT44257.1 GFDL01010253 JAV24792.1 DS232020 EDS32140.1 MCFN01000062 OXB66522.1 AWGT02000046 OXB80629.1 PPHD01078481 POI20758.1 AAQR03116380 AK005756 BC028657 AADN05000150 AABR07051243 CH474001 EDL93563.1 GFFW01004515 JAV40273.1 GBHO01031190 GBHO01031189 GBHO01031188 GBHO01031184 GBHO01003880 JAG12414.1 JAG12415.1 JAG12416.1 JAG12420.1 JAG39724.1 GAPW01006071 JAC07527.1 AGTP01090703 AMQN01011090 KB308677 ELT97046.1 BC108196 UYRR01034292 VDK60731.1 GDAI01001059 JAI16544.1
SOQ38909.1 KZ150118 PZC73236.1 NWSH01001095 PCG72639.1 GAIX01006190 JAA86370.1 JTDY01003397 KOB69620.1 AGBW02010179 OWR49018.1 KQ459465 KPJ00309.1 KQ460393 KPJ15379.1 LADJ01008100 KPJ20915.1 GEBQ01018260 GEBQ01011159 JAT21717.1 JAT28818.1 GGFL01006614 MBW70792.1 GECL01000722 JAP05402.1 GECZ01001160 JAS68609.1 GGFK01012324 MBW45645.1 GBGD01003493 JAC85396.1 GGFM01005820 MBW26571.1 GGFJ01010752 MBW59893.1 UFQT01000991 SSX28326.1 AXCM01002444 GGFK01012207 MBW45528.1 APCN01002026 AXCN02001153 GBRD01001291 JAG64530.1 GBHO01031187 GBHO01031186 GBHO01031185 GBHO01031183 GBHO01003881 GBHO01003879 GDHC01015863 GDHC01005617 JAG12417.1 JAG12418.1 JAG12419.1 JAG12421.1 JAG39723.1 JAG39725.1 JAQ02766.1 JAQ13012.1 JRES01000378 KNC31832.1 CM000070 EIM52294.1 S77099 GDKW01003164 JAI53431.1 GAHY01000908 JAA76602.1 GBBI01001275 JAC17437.1 AAAB01008987 EAA01434.5 GFTR01002098 JAW14328.1 GGFL01004544 MBW68722.1 GECU01031236 JAS76470.1 AYCK01016959 AYCK01026587 CH964272 EDW83917.1 CH479186 EDW39303.1 FJ821034 ACN94747.1 GANO01003398 JAB56473.1 GBYX01173258 JAO87841.1 GCES01123887 JAQ62435.1 OUUW01000005 SPP80941.1 GCES01011505 JAR74818.1 KZ288256 PBC30636.1 JXUM01144562 KQ569817 KXJ68510.1 JH002744 RAZU01000235 EGW14296.1 RLQ63888.1 CP012526 ALC46262.1 CH477301 EAT44257.1 GFDL01010253 JAV24792.1 DS232020 EDS32140.1 MCFN01000062 OXB66522.1 AWGT02000046 OXB80629.1 PPHD01078481 POI20758.1 AAQR03116380 AK005756 BC028657 AADN05000150 AABR07051243 CH474001 EDL93563.1 GFFW01004515 JAV40273.1 GBHO01031190 GBHO01031189 GBHO01031188 GBHO01031184 GBHO01003880 JAG12414.1 JAG12415.1 JAG12416.1 JAG12420.1 JAG39724.1 GAPW01006071 JAC07527.1 AGTP01090703 AMQN01011090 KB308677 ELT97046.1 BC108196 UYRR01034292 VDK60731.1 GDAI01001059 JAI16544.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000075900 UP000095301 UP000075920 UP000076408 UP000075901 UP000069272 UP000075883 UP000005203 UP000075840 UP000261500 UP000075886 UP000075885 UP000075884 UP000075880 UP000037069 UP000001819 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075881 UP000261480 UP000028760 UP000007798 UP000008744 UP000268350 UP000002852 UP000242457 UP000069940 UP000249989 UP000001075 UP000273346 UP000002280 UP000092553 UP000008820 UP000002320 UP000091820 UP000198323 UP000198419 UP000005225 UP000000589 UP000078200 UP000000539 UP000002494 UP000261520 UP000005215 UP000233160 UP000007635 UP000265180 UP000001038 UP000261560 UP000014760 UP000009136 UP000036680 UP000189706 UP000265200
UP000075900 UP000095301 UP000075920 UP000076408 UP000075901 UP000069272 UP000075883 UP000005203 UP000075840 UP000261500 UP000075886 UP000075885 UP000075884 UP000075880 UP000037069 UP000001819 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075881 UP000261480 UP000028760 UP000007798 UP000008744 UP000268350 UP000002852 UP000242457 UP000069940 UP000249989 UP000001075 UP000273346 UP000002280 UP000092553 UP000008820 UP000002320 UP000091820 UP000198323 UP000198419 UP000005225 UP000000589 UP000078200 UP000000539 UP000002494 UP000261520 UP000005215 UP000233160 UP000007635 UP000265180 UP000001038 UP000261560 UP000014760 UP000009136 UP000036680 UP000189706 UP000265200
Pfam
PF05005 Ocnus
Gene 3D
ProteinModelPortal
Q2F5N5
H9JVD0
A0A1E1VX92
A0A2H1VDJ4
A0A2W1BK75
A0A2A4JLE7
+ More
S4PGJ4 A0A0L7L2G8 A0A212F5L1 A0A194Q4N4 A0A0N1PGL2 A0A0N1PI35 A0A182R4T0 A0A1I8NB48 A0A1B6LYP8 A0A2M4CZN2 A0A0V0GDK6 A0A182VWL1 A0A182YHG7 A0A182T9L2 A0A1B6H1Q8 A0A2M4AY21 A0A069DP21 A0A2M3ZDK6 A0A182FNZ3 A0A2M4C3L3 A0A336MDK8 A0A182MKI6 A0A088ACD4 A0A2M4AY66 A0A182HZZ2 A0A3B3U8L0 A0A182Q8C2 A0A182P575 A0A182N5U6 A0A182ILD5 A0A0K8THC0 A0A0A9ZD48 A0A0L0CHZ4 I5AMV9 P54364 A0A0P4VLC5 R4FMN8 A0A023F860 A0A182V6Q2 A0A182TJU1 A0A182LR35 A0A182X0A2 Q7PUI9 A0A224XWS2 A0A2M4CTP6 A0A1B6HP61 A0A1S4F7C0 A0A182JVI0 A0A3B3WX09 A0A087X7T5 A0A087X5G2 B4NJJ3 B4GNH7 D0QWR6 U5EVH9 A0A0S7LEG5 A0A146R1V4 A0A3B0K7G7 A0A147A8G3 M3ZLH2 A0A2A3EHH3 A0A182G4V8 G3IHB0 F6WVK9 A0A0M4F3P7 Q17D02 A0A1Q3FBA0 B0WP95 A0A1A9WT60 A0A226NGC1 A0A226PMI4 A0A2P4S9G9 H0XUK7 Q9DAK9 A0A1A9VDH0 A0A1D5PWG8 D3ZP47 A0A3B4ANQ6 A0A250Y9L6 A0A3B4AME5 A0A0A9X5P1 A0A023EDZ0 I3NEJ4 A0A2K6EIY3 G3Q237 A0A3P9K6C4 A0A3B3H9F4 A0A3B3C695 R7U055 Q32PA4 A0A0M3KB29 A0A1U7R1F2 A0A3P9JMV2 A0A0K8TQA2
S4PGJ4 A0A0L7L2G8 A0A212F5L1 A0A194Q4N4 A0A0N1PGL2 A0A0N1PI35 A0A182R4T0 A0A1I8NB48 A0A1B6LYP8 A0A2M4CZN2 A0A0V0GDK6 A0A182VWL1 A0A182YHG7 A0A182T9L2 A0A1B6H1Q8 A0A2M4AY21 A0A069DP21 A0A2M3ZDK6 A0A182FNZ3 A0A2M4C3L3 A0A336MDK8 A0A182MKI6 A0A088ACD4 A0A2M4AY66 A0A182HZZ2 A0A3B3U8L0 A0A182Q8C2 A0A182P575 A0A182N5U6 A0A182ILD5 A0A0K8THC0 A0A0A9ZD48 A0A0L0CHZ4 I5AMV9 P54364 A0A0P4VLC5 R4FMN8 A0A023F860 A0A182V6Q2 A0A182TJU1 A0A182LR35 A0A182X0A2 Q7PUI9 A0A224XWS2 A0A2M4CTP6 A0A1B6HP61 A0A1S4F7C0 A0A182JVI0 A0A3B3WX09 A0A087X7T5 A0A087X5G2 B4NJJ3 B4GNH7 D0QWR6 U5EVH9 A0A0S7LEG5 A0A146R1V4 A0A3B0K7G7 A0A147A8G3 M3ZLH2 A0A2A3EHH3 A0A182G4V8 G3IHB0 F6WVK9 A0A0M4F3P7 Q17D02 A0A1Q3FBA0 B0WP95 A0A1A9WT60 A0A226NGC1 A0A226PMI4 A0A2P4S9G9 H0XUK7 Q9DAK9 A0A1A9VDH0 A0A1D5PWG8 D3ZP47 A0A3B4ANQ6 A0A250Y9L6 A0A3B4AME5 A0A0A9X5P1 A0A023EDZ0 I3NEJ4 A0A2K6EIY3 G3Q237 A0A3P9K6C4 A0A3B3H9F4 A0A3B3C695 R7U055 Q32PA4 A0A0M3KB29 A0A1U7R1F2 A0A3P9JMV2 A0A0K8TQA2
PDB
2NMM
E-value=5.35572e-26,
Score=286
Ontologies
KEGG
GO
PANTHER
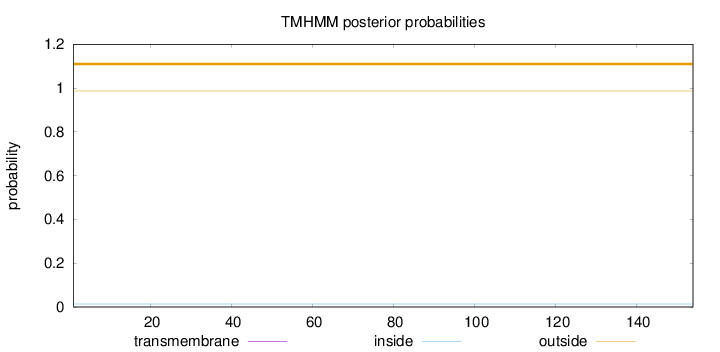
Topology
Subcellular location
Cytoplasm
Length:
154
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.01367
outside
1 - 154
Population Genetic Test Statistics
Pi
214.336827
Theta
161.755598
Tajima's D
0.973545
CLR
0.007425
CSRT
0.654167291635418
Interpretation
Uncertain
