Pre Gene Modal
BGIBMGA013492
Annotation
PREDICTED:_carboxypeptidase_N_subunit_2-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.185 Extracellular Reliability : 1.246 Nuclear Reliability : 1.154
Sequence
CDS
ATGGCTCTCAAAACAAGCTTAGTGTTCTGTCTCATCGCAACGGGTCTTAGCTACGCTTTCAACGGGGACAGCTTCGAGCTGGAGTGCCCGGACGAGTGCGACTGCCACTACTTCAGAATCAACTGGGTCACGGACTGTTCAGAGAGTAACCTCACCGAAGTGCCATATGATGAGCTCAGCCTCAGCGTGTATATTCTAGATTTGAACGGCAACAACATTACAACCCTCAAGCCCTTCCCGAACGATATCAAGATGAGGAGATTGCAAATAGCTGATAATCGACTGACCAGAGTCGAACGTGAAGCGTTCAAGGGCCTGGAATACTTGATTGATATTGATTTGTCTGGAAACAACATCAGTTATGTGGACCCCGAGGCGTTCTTGGATTCCCGCGGTTTGCTAAACGTGGAGCTTCAAGACAATCCAATAGGCAACGTCGAAGGCCCCTTCCTCGTCTCTCCAACGTTGCAGTATCTCGATTTAAGTAACTGCAATATAACCTCCATCAATTCTCAATTCTTTGACAATATCACAACCCTTATGACAGTAGACCTCTCCAATAATCCTCTGAAAAATTTAGCAACCGGCGTTTTCGATATCTTAACAAGCATAGAAACTTTGAAACTCAACGACTGCAAGCTCTCGAACATTTCAGAAGTTTTATTCGCTAATGCTGCACACCTGAAGAATTTGGAATTGGCCGGAAACTATCTAAAGCACACAGATTGGTCTGAAGTACTCGGAAAATTGGTCAGACTAGAATATCTGAATTTGAGGAAAACTGGTATAACATACCTTTCAGAAGACACATTTTCGAATTGTACGTATTTGGTGACGTTAGTACTTGCAGATAATGAGCTGCATGATTTAGACGTCGCTGCTACGTTGGGCAGTAGCGTGAATCATTTAGAATTGCTCGATTTATCCAATTGCCGCATCCAGGGCCCTCTGTCCGGCGAAGCGTTCGCTAATGCGACCAGATTGAAAAGTCTTTATCTTTCTGGTAATCCACTATTTGCGGCAGACTTACAAGAAGCGCTCGCTCCGCTACCGAAATTGGAAAGGTTGTTCTTAAGCAATTGTGGCCTACGCAATCTTCCAGATAACTTTAATGTTTTCGACAATCTTTTAGAACTAGACATATCGCATAATCCTCTGAATAATGTTTTCACGAGACTTTTAGCTCCATTAGAAAAATTAGAATACTTGAATATGGGTCACAGTAATTTATCGTATATTGGGCCAGATTCGTTCGCCAAGATGACCTCGATGAAAAGATTAGTTCTCTCTGGAAACGACTTGCTGTCCCTAGAAGCCGGTCTCTTCGGTAATCTGACGCAACTAACTACGTTAGAATTAGAAATGTGTGGCTTAAAACGGCCCTTAAACGCTAACGTATTTTTTAAAAACCTAACTTATATGGACTTGAGAGAAATAAAACTAGGAAGTAATCCTTTGGTTATTCCCGATAAAGGACCCGTGTTCCCGAAACAGCTGTCGCAAGTAACGACGCTCGATTTAAGTAATTGTAATATAACATCACTCAAAGTTGATGCTTTCAAAAACACAATAAACTTAACTGAACTGAATCTGTCAGGCAATAGATTACGATCTAACGATGGAAATTTGGCATTTTTGGAAATCTTAAAGCATCTAGAGAAAATAAATCTAAGCTATAATAATTTAACGACAATCGATCCGCAAATTTTCACGCACAATCCGAAATTACATTCTTTGAACCTGCTAGGGAATCCATTTATTTGCGACTGCAAAATCGCTGAGATGTGGGACTGGGCTAACATGATAAAGGGTGACTTAGACGTGCTCGTTGGGGCCAAGAGTGCTGAAAAGGATATCGTTGTAAAAGGAAATAAGAAGAAAAAATATCTATACTGTCATTATAATGAAACTCAACTTCGAAATGCGAGTTTAATGTTAAACAAGACAGTACCAGGCAGAAGACCGTTCATAAAACCAAGAGAACTTATTTACGCTAACAGGACTTGGGCAAAATATGTGCGGGAATCTGGATGCGAGCCCGTCGTGAAAATATTGAGGCCCATAGCTTTGGTAGCAGAGATATTTGATTTCACGAACGGACCTAAAATACCTACCGGCGTTGTAATAGTTGCATTAATAGCGATTATCCTTGGTTTAGCCACGCTTATTATGACGATAAGGTTGTTCAGGAGAAACAACACCACGGAAATAGACTCTGAAACTAGCAGAAAATCAAGATAG
Protein
MALKTSLVFCLIATGLSYAFNGDSFELECPDECDCHYFRINWVTDCSESNLTEVPYDELSLSVYILDLNGNNITTLKPFPNDIKMRRLQIADNRLTRVEREAFKGLEYLIDIDLSGNNISYVDPEAFLDSRGLLNVELQDNPIGNVEGPFLVSPTLQYLDLSNCNITSINSQFFDNITTLMTVDLSNNPLKNLATGVFDILTSIETLKLNDCKLSNISEVLFANAAHLKNLELAGNYLKHTDWSEVLGKLVRLEYLNLRKTGITYLSEDTFSNCTYLVTLVLADNELHDLDVAATLGSSVNHLELLDLSNCRIQGPLSGEAFANATRLKSLYLSGNPLFAADLQEALAPLPKLERLFLSNCGLRNLPDNFNVFDNLLELDISHNPLNNVFTRLLAPLEKLEYLNMGHSNLSYIGPDSFAKMTSMKRLVLSGNDLLSLEAGLFGNLTQLTTLELEMCGLKRPLNANVFFKNLTYMDLREIKLGSNPLVIPDKGPVFPKQLSQVTTLDLSNCNITSLKVDAFKNTINLTELNLSGNRLRSNDGNLAFLEILKHLEKINLSYNNLTTIDPQIFTHNPKLHSLNLLGNPFICDCKIAEMWDWANMIKGDLDVLVGAKSAEKDIVVKGNKKKKYLYCHYNETQLRNASLMLNKTVPGRRPFIKPRELIYANRTWAKYVRESGCEPVVKILRPIALVAEIFDFTNGPKIPTGVVIVALIAIILGLATLIMTIRLFRRNNTTEIDSETSRKSR
Summary
Uniprot
H9JVC9
A0A2W1BDD5
A0A2A4JML5
A0A2H1X3D7
A0A0L7L2W0
A0A194Q443
+ More
A0A0N0PCZ7 A0A212F5K3 D7EK56 N6UHC2 A0A1W4WQ31 A0A1W4WZS9 A0A0L7QZ24 A0A1W6EW78 A0A2S2PVZ6 J9JKN4 A0A2H8TZN6 A0A232FMK1 A0A2S2PKZ3 A0A2J7QKT0 A0A088AQB0 A0A0N0BEA7 K7JUD3 A0A158P3Z7 A0A195BL10 A0A2A3EIA9 A0A195C2V3 A0A151JM50 F4WKR9 V9IK06 A0A151JZ18 A0A3L8DCN1 A0A067RCH3 E9J8X5 E2C619 A0A2P8YRQ7 V5GZU2 A0A0C9RWG7 N0A0P9 E2AZ31 A0A3Q0JJN8 A0A286JZ77 A0A146KNW1 A0A0K8S5S2 A0A0A9WPE7 A0A0J7L774 A0A1B6CHW6 A0A1B6FJK1 A0A1B6IQ30 A0A026WJV5 A0A154PL32 A0A224X852 A0A2R7WIJ4 T1HL24 A0A2P8ZN27 A0A2P6SHC1 A0A177AZC7 A0A2P6SGN3 E2BAJ0 A0A2P8YF33 A0A067RI05 A0A146ZXB2 A0A1W4XCF5 A0A2P2HXK5 A0A0D2WUF4 A0A182HEB3 A0A3B3YP01 A0A182V6Y3 A0A182XUD2 J9KX32 Q5TWN6 A0A182UJW5 A0A3B4T9T0 A0A182MDJ2 B4JS99 A0A1S3IHZ9 A0A084WJS6 A0A151J9Q9 A0A182L386 B0WH32 A0A182IIJ9 A0A182WL04 F4WEC8 A0A226EY20 A0A1S3IYX6 A0A151WEM2 A0A224XKI5 A0A1B0A582 A0A1A9VPY0 A0A195FGY5 A0A1B0GE40 A0A195BEP1 B3LYV9 A0A0P8YPB8 A0A182RMZ2
A0A0N0PCZ7 A0A212F5K3 D7EK56 N6UHC2 A0A1W4WQ31 A0A1W4WZS9 A0A0L7QZ24 A0A1W6EW78 A0A2S2PVZ6 J9JKN4 A0A2H8TZN6 A0A232FMK1 A0A2S2PKZ3 A0A2J7QKT0 A0A088AQB0 A0A0N0BEA7 K7JUD3 A0A158P3Z7 A0A195BL10 A0A2A3EIA9 A0A195C2V3 A0A151JM50 F4WKR9 V9IK06 A0A151JZ18 A0A3L8DCN1 A0A067RCH3 E9J8X5 E2C619 A0A2P8YRQ7 V5GZU2 A0A0C9RWG7 N0A0P9 E2AZ31 A0A3Q0JJN8 A0A286JZ77 A0A146KNW1 A0A0K8S5S2 A0A0A9WPE7 A0A0J7L774 A0A1B6CHW6 A0A1B6FJK1 A0A1B6IQ30 A0A026WJV5 A0A154PL32 A0A224X852 A0A2R7WIJ4 T1HL24 A0A2P8ZN27 A0A2P6SHC1 A0A177AZC7 A0A2P6SGN3 E2BAJ0 A0A2P8YF33 A0A067RI05 A0A146ZXB2 A0A1W4XCF5 A0A2P2HXK5 A0A0D2WUF4 A0A182HEB3 A0A3B3YP01 A0A182V6Y3 A0A182XUD2 J9KX32 Q5TWN6 A0A182UJW5 A0A3B4T9T0 A0A182MDJ2 B4JS99 A0A1S3IHZ9 A0A084WJS6 A0A151J9Q9 A0A182L386 B0WH32 A0A182IIJ9 A0A182WL04 F4WEC8 A0A226EY20 A0A1S3IYX6 A0A151WEM2 A0A224XKI5 A0A1B0A582 A0A1A9VPY0 A0A195FGY5 A0A1B0GE40 A0A195BEP1 B3LYV9 A0A0P8YPB8 A0A182RMZ2
Pubmed
EMBL
BABH01036055
KZ150118
PZC73239.1
NWSH01001095
PCG72642.1
ODYU01013075
+ More
SOQ59756.1 JTDY01003397 KOB69619.1 KQ459465 KPJ00312.1 KQ460393 KPJ15377.1 AGBW02010179 OWR49015.1 DS497731 EFA13011.1 APGK01020354 KB740193 KB631669 ENN81115.1 ERL85229.1 KQ414685 KOC63826.1 KY563563 ARK19972.1 GGMS01000337 MBY69540.1 ABLF02035549 GFXV01006813 MBW18618.1 NNAY01000046 OXU31587.1 GGMR01017490 MBY30109.1 NEVH01013256 PNF29192.1 KQ435840 KOX71372.1 ADTU01008653 ADTU01008654 KQ976453 KYM85367.1 KZ288232 PBC31450.1 KQ978317 KYM95167.1 KQ978949 KYN27380.1 GL888206 EGI65161.1 JR049499 AEY60982.1 KQ981427 KYN41762.1 QOIP01000010 RLU17902.1 KK852549 KDR21566.1 GL769162 EFZ10733.1 GL452895 EFN76624.1 PYGN01000405 PSN46923.1 GALX01002408 JAB66058.1 GBYB01013330 JAG83097.1 KC355193 AGK40893.1 GL444078 EFN61305.1 KU866525 AMQ10343.1 GDHC01021254 JAP97374.1 GBRD01017757 JAG48070.1 GBHO01033262 JAG10342.1 LBMM01000409 KMQ98428.1 GEDC01024269 JAS13029.1 GECZ01019410 JAS50359.1 GECU01018677 JAS89029.1 KK107167 EZA56290.1 KQ434936 KZC11940.1 GFTR01007900 JAW08526.1 KK854726 PTY18215.1 ACPB03005246 PYGN01000013 PSN57880.1 PDCK01000039 PRQ58036.1 LWCA01000718 OAF67220.1 PRQ57855.1 GL446759 EFN87284.1 PYGN01000649 PSN42843.1 KK852461 KDR23437.1 GCES01015363 JAR70960.1 IACF01000747 LAB66503.1 KE346371 KJE96300.1 JXUM01131322 JXUM01131323 KQ567697 KXJ69314.1 ABLF02007606 ABLF02009963 ABLF02009965 AAAB01008807 EAL41884.2 AXCM01009741 CH916373 EDV94639.1 ATLV01024053 ATLV01024054 KE525348 KFB50470.1 KQ979388 KYN21757.1 DS231932 EDS27483.1 APCN01001490 GL888102 EGI67566.1 LNIX01000001 OXA61496.1 KQ983238 KYQ46304.1 GFTR01007897 JAW08529.1 KQ981606 KYN39631.1 CCAG010012086 CCAG010012087 KQ976509 KYM82630.1 CH902617 EDV44075.2 KPU80609.1
SOQ59756.1 JTDY01003397 KOB69619.1 KQ459465 KPJ00312.1 KQ460393 KPJ15377.1 AGBW02010179 OWR49015.1 DS497731 EFA13011.1 APGK01020354 KB740193 KB631669 ENN81115.1 ERL85229.1 KQ414685 KOC63826.1 KY563563 ARK19972.1 GGMS01000337 MBY69540.1 ABLF02035549 GFXV01006813 MBW18618.1 NNAY01000046 OXU31587.1 GGMR01017490 MBY30109.1 NEVH01013256 PNF29192.1 KQ435840 KOX71372.1 ADTU01008653 ADTU01008654 KQ976453 KYM85367.1 KZ288232 PBC31450.1 KQ978317 KYM95167.1 KQ978949 KYN27380.1 GL888206 EGI65161.1 JR049499 AEY60982.1 KQ981427 KYN41762.1 QOIP01000010 RLU17902.1 KK852549 KDR21566.1 GL769162 EFZ10733.1 GL452895 EFN76624.1 PYGN01000405 PSN46923.1 GALX01002408 JAB66058.1 GBYB01013330 JAG83097.1 KC355193 AGK40893.1 GL444078 EFN61305.1 KU866525 AMQ10343.1 GDHC01021254 JAP97374.1 GBRD01017757 JAG48070.1 GBHO01033262 JAG10342.1 LBMM01000409 KMQ98428.1 GEDC01024269 JAS13029.1 GECZ01019410 JAS50359.1 GECU01018677 JAS89029.1 KK107167 EZA56290.1 KQ434936 KZC11940.1 GFTR01007900 JAW08526.1 KK854726 PTY18215.1 ACPB03005246 PYGN01000013 PSN57880.1 PDCK01000039 PRQ58036.1 LWCA01000718 OAF67220.1 PRQ57855.1 GL446759 EFN87284.1 PYGN01000649 PSN42843.1 KK852461 KDR23437.1 GCES01015363 JAR70960.1 IACF01000747 LAB66503.1 KE346371 KJE96300.1 JXUM01131322 JXUM01131323 KQ567697 KXJ69314.1 ABLF02007606 ABLF02009963 ABLF02009965 AAAB01008807 EAL41884.2 AXCM01009741 CH916373 EDV94639.1 ATLV01024053 ATLV01024054 KE525348 KFB50470.1 KQ979388 KYN21757.1 DS231932 EDS27483.1 APCN01001490 GL888102 EGI67566.1 LNIX01000001 OXA61496.1 KQ983238 KYQ46304.1 GFTR01007897 JAW08529.1 KQ981606 KYN39631.1 CCAG010012086 CCAG010012087 KQ976509 KYM82630.1 CH902617 EDV44075.2 KPU80609.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000007266 UP000019118 UP000030742 UP000192223 UP000053825 UP000007819 UP000215335 UP000235965 UP000005203 UP000053105 UP000002358 UP000005205 UP000078540 UP000242457 UP000078542 UP000078492 UP000007755 UP000078541 UP000279307 UP000027135 UP000008237 UP000245037 UP000000311 UP000079169 UP000036403 UP000053097 UP000076502 UP000015103 UP000238479 UP000265000 UP000008743 UP000069940 UP000249989 UP000261480 UP000075903 UP000076407 UP000007062 UP000075902 UP000261420 UP000075883 UP000001070 UP000085678 UP000030765 UP000075882 UP000002320 UP000075840 UP000075920 UP000198287 UP000075809 UP000092445 UP000078200 UP000092444 UP000007801 UP000075900
UP000007266 UP000019118 UP000030742 UP000192223 UP000053825 UP000007819 UP000215335 UP000235965 UP000005203 UP000053105 UP000002358 UP000005205 UP000078540 UP000242457 UP000078542 UP000078492 UP000007755 UP000078541 UP000279307 UP000027135 UP000008237 UP000245037 UP000000311 UP000079169 UP000036403 UP000053097 UP000076502 UP000015103 UP000238479 UP000265000 UP000008743 UP000069940 UP000249989 UP000261480 UP000075903 UP000076407 UP000007062 UP000075902 UP000261420 UP000075883 UP000001070 UP000085678 UP000030765 UP000075882 UP000002320 UP000075840 UP000075920 UP000198287 UP000075809 UP000092445 UP000078200 UP000092444 UP000007801 UP000075900
PRIDE
Pfam
Interpro
IPR032675
LRR_dom_sf
+ More
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR026906 LRR_5
IPR025875 Leu-rich_rpt_4
IPR000372 LRRNT
IPR000073 AB_hydrolase_1
IPR000952 AB_hydrolase_4_CS
IPR029058 AB_hydrolase
IPR013210 LRR_N_plant-typ
IPR008855 TRAP-delta
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR000483 Cys-rich_flank_reg_C
IPR036116 FN3_sf
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR026906 LRR_5
IPR025875 Leu-rich_rpt_4
IPR000372 LRRNT
IPR000073 AB_hydrolase_1
IPR000952 AB_hydrolase_4_CS
IPR029058 AB_hydrolase
IPR013210 LRR_N_plant-typ
IPR008855 TRAP-delta
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR000483 Cys-rich_flank_reg_C
IPR036116 FN3_sf
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
SUPFAM
Gene 3D
ProteinModelPortal
H9JVC9
A0A2W1BDD5
A0A2A4JML5
A0A2H1X3D7
A0A0L7L2W0
A0A194Q443
+ More
A0A0N0PCZ7 A0A212F5K3 D7EK56 N6UHC2 A0A1W4WQ31 A0A1W4WZS9 A0A0L7QZ24 A0A1W6EW78 A0A2S2PVZ6 J9JKN4 A0A2H8TZN6 A0A232FMK1 A0A2S2PKZ3 A0A2J7QKT0 A0A088AQB0 A0A0N0BEA7 K7JUD3 A0A158P3Z7 A0A195BL10 A0A2A3EIA9 A0A195C2V3 A0A151JM50 F4WKR9 V9IK06 A0A151JZ18 A0A3L8DCN1 A0A067RCH3 E9J8X5 E2C619 A0A2P8YRQ7 V5GZU2 A0A0C9RWG7 N0A0P9 E2AZ31 A0A3Q0JJN8 A0A286JZ77 A0A146KNW1 A0A0K8S5S2 A0A0A9WPE7 A0A0J7L774 A0A1B6CHW6 A0A1B6FJK1 A0A1B6IQ30 A0A026WJV5 A0A154PL32 A0A224X852 A0A2R7WIJ4 T1HL24 A0A2P8ZN27 A0A2P6SHC1 A0A177AZC7 A0A2P6SGN3 E2BAJ0 A0A2P8YF33 A0A067RI05 A0A146ZXB2 A0A1W4XCF5 A0A2P2HXK5 A0A0D2WUF4 A0A182HEB3 A0A3B3YP01 A0A182V6Y3 A0A182XUD2 J9KX32 Q5TWN6 A0A182UJW5 A0A3B4T9T0 A0A182MDJ2 B4JS99 A0A1S3IHZ9 A0A084WJS6 A0A151J9Q9 A0A182L386 B0WH32 A0A182IIJ9 A0A182WL04 F4WEC8 A0A226EY20 A0A1S3IYX6 A0A151WEM2 A0A224XKI5 A0A1B0A582 A0A1A9VPY0 A0A195FGY5 A0A1B0GE40 A0A195BEP1 B3LYV9 A0A0P8YPB8 A0A182RMZ2
A0A0N0PCZ7 A0A212F5K3 D7EK56 N6UHC2 A0A1W4WQ31 A0A1W4WZS9 A0A0L7QZ24 A0A1W6EW78 A0A2S2PVZ6 J9JKN4 A0A2H8TZN6 A0A232FMK1 A0A2S2PKZ3 A0A2J7QKT0 A0A088AQB0 A0A0N0BEA7 K7JUD3 A0A158P3Z7 A0A195BL10 A0A2A3EIA9 A0A195C2V3 A0A151JM50 F4WKR9 V9IK06 A0A151JZ18 A0A3L8DCN1 A0A067RCH3 E9J8X5 E2C619 A0A2P8YRQ7 V5GZU2 A0A0C9RWG7 N0A0P9 E2AZ31 A0A3Q0JJN8 A0A286JZ77 A0A146KNW1 A0A0K8S5S2 A0A0A9WPE7 A0A0J7L774 A0A1B6CHW6 A0A1B6FJK1 A0A1B6IQ30 A0A026WJV5 A0A154PL32 A0A224X852 A0A2R7WIJ4 T1HL24 A0A2P8ZN27 A0A2P6SHC1 A0A177AZC7 A0A2P6SGN3 E2BAJ0 A0A2P8YF33 A0A067RI05 A0A146ZXB2 A0A1W4XCF5 A0A2P2HXK5 A0A0D2WUF4 A0A182HEB3 A0A3B3YP01 A0A182V6Y3 A0A182XUD2 J9KX32 Q5TWN6 A0A182UJW5 A0A3B4T9T0 A0A182MDJ2 B4JS99 A0A1S3IHZ9 A0A084WJS6 A0A151J9Q9 A0A182L386 B0WH32 A0A182IIJ9 A0A182WL04 F4WEC8 A0A226EY20 A0A1S3IYX6 A0A151WEM2 A0A224XKI5 A0A1B0A582 A0A1A9VPY0 A0A195FGY5 A0A1B0GE40 A0A195BEP1 B3LYV9 A0A0P8YPB8 A0A182RMZ2
PDB
4KNG
E-value=5.45271e-23,
Score=269
Ontologies
GO
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.998727
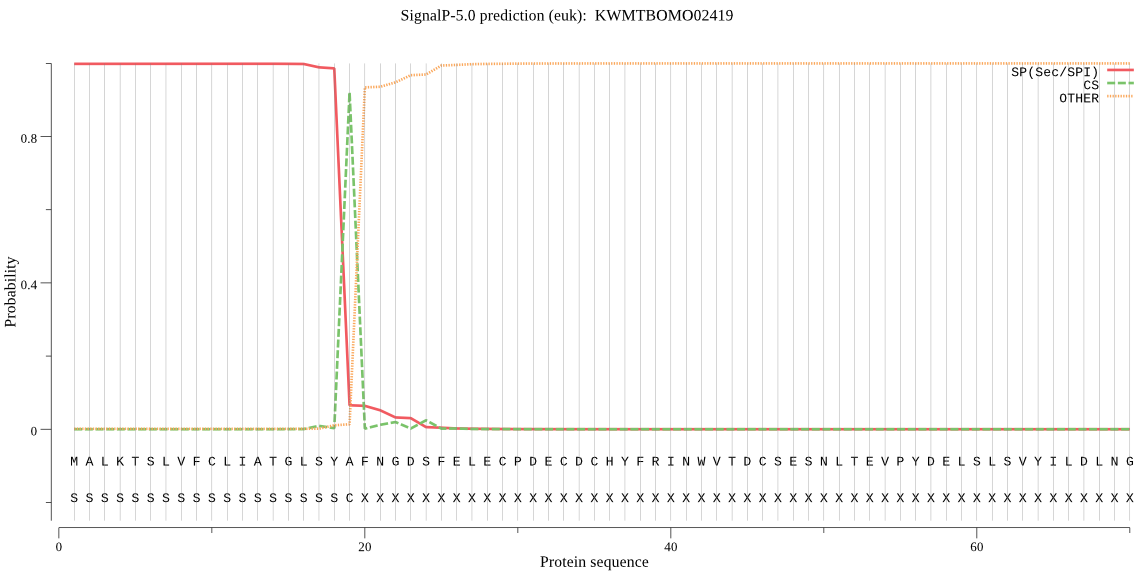
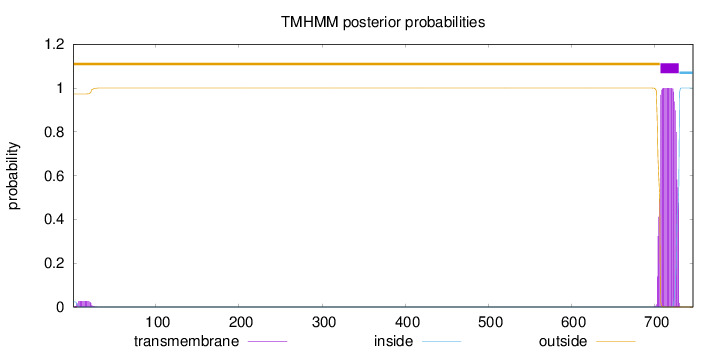
Length:
746
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.05748
Exp number, first 60 AAs:
0.47626
Total prob of N-in:
0.02665
outside
1 - 706
TMhelix
707 - 729
inside
730 - 746
Population Genetic Test Statistics
Pi
178.84422
Theta
172.909722
Tajima's D
0.189124
CLR
0.442672
CSRT
0.428628568571571
Interpretation
Uncertain
