Gene
KWMTBOMO02407
Pre Gene Modal
BGIBMGA012097
Annotation
PREDICTED:_uncharacterized_protein_LOC101739096_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.984
Sequence
CDS
ATGTTCTGGCTGGGCTTCGTGGGTCAAACGGTGGTGGCCATCTCGCAGGTGTTCATCCTAAACGTACCTCCGCGGCTGGCGGCCGTGTGGTTTGGGGCCGATCAAGTGTCGTCCGCTTGCAGTATCGGCGTTTTCGGTAATCAGTTAGGCGTTGCTCTCGGATTTTTGCTGCCTCCGATGCTGGTTAAATCGACCGGTACTATGGAACAGATCGGTCATGATTTCCAGCTCATGTTCTACCTGGTCGCCGGCTTCACCAGCGTCCTTTTCGTCTTCATACTGCTATTTTTCAAAGCGGCACCACCAACCCCGCCATCAGCGGCTGCTGATCTTGGCTCAACTATCGACTCTAACTTCTTGCAATCGATCAAGACGCTACTAATGAACAGAAACTACGTGTTGCTGCTTATCTCGTATGGCCTGAACGTCGGTACGTTCTATGCTATCTCGACTCTGCTCAATCGAGTCATCCTGACTTATTATCCGGGCGCGAGTGAAGACGCTGGACGAATTGGGCTTGTAATCGTGGTCGCGGGTATGGCCGGCTCCGTCGTTTGTGGTCTGATCCTCGACAAGACGCATCGGTTCAAGGAGACAACCCTGGCGGTTTACACGTCCTCTGTCATCGGCATGATCATATTTACCTTCACATTGGATTGCGGATTGATCGGCGTGGTTTATTTGAGCAGCATAATACTTGGATTCTTCATGACCGGCTACTTGCCTGTTGGCTTCGAGTTCGCTTCCGAAGTGACGTACCCTGAACCAGAAGGGACCACCTCCGGAATACTGAACGCTGTTGTACAGGTGTTCGGTATACTGACGACGCTGCTGTACGGCTGGATGCTGGACGAGGTCGGCGACCGCTGGGCCAACCTCACGCTGTGCGCGCTGCTGGCGCTGGGCGCCGCCGTCACCGCCGCCATCCGCTCGGACCTGCGGCGCCAGGCGGCACAGAACAAGGCCTGA
Protein
MFWLGFVGQTVVAISQVFILNVPPRLAAVWFGADQVSSACSIGVFGNQLGVALGFLLPPMLVKSTGTMEQIGHDFQLMFYLVAGFTSVLFVFILLFFKAAPPTPPSAAADLGSTIDSNFLQSIKTLLMNRNYVLLLISYGLNVGTFYAISTLLNRVILTYYPGASEDAGRIGLVIVVAGMAGSVVCGLILDKTHRFKETTLAVYTSSVIGMIIFTFTLDCGLIGVVYLSSIILGFFMTGYLPVGFEFASEVTYPEPEGTTSGILNAVVQVFGILTTLLYGWMLDEVGDRWANLTLCALLALGAAVTAAIRSDLRRQAAQNKA
Summary
Uniprot
A0A212F6Y2
A0A2A4IXF8
A0A194Q455
A0A2W1BEJ5
A0A0N1PIB1
H9JRD7
+ More
A0A2H1VM50 U5ERN6 A0A336KTF1 A0A1Q3FJ03 Q17AU9 A0A1Q3FJ69 B0W3Q5 Q7PNM1 A0A1I8JVP8 A0A067REN7 A0A1J1HXI6 A0A2M4BLW0 D6WNC0 A0A084WE82 W5J8W1 A0A1Y1LT21 A0A2J7R2R2 A0A2M3Z4T4 A0A1Y1LP75 A0A1Y1LR45 A0A2J7R2Q8 A0A0L0C1H5 A0A154P3V2 A0A1I8MDL9 T1P881 B3MHR6 A0A0M4EKR8 B4H8I7 A0A0L7LGQ7 Q28YK0 B4QEP7 B4HR44 Q7K1D7 A0A3B0JPZ5 B3N9W4 A0A1W4VBL3 B4P1S4 B4LJG3 B4J7Y9 A0A1I8PJP5 K7J1E9 B4KPQ6 A0A182QSK1 A0A1B6KB51 A0A1B6E568 A0A1B6ECM9 A0A1B6H316 A0A1B6EIH7 A0A1B6CJ43 B4MIP5 A0A1B6DT76 E9HG13 A0A1B6EH14 A0A232FET5 V5G4X6 A0A0A1XE15 A0A0L7R1Y5 A0A026WQE7 A0A2S2Q2E4 A0A0N8CTS5 A0A310SIR3 A0A0P5V8Q8 A0A0P5Z5F3 A0A0P5E5C2 E9H209 A0A0C9RFK2 C4WRM7 A0A0P6DT70 A0A2S2PT36 A0A1A9ZYG6 A0A0C9R6F8 A0A088A629 A0A0P5GFG3 A0A0P4XNY6 A0A0P6HJA0 V9I6K4 J9JQF9 V9I6M3 W8BIC4 A0A2A3EIP9 A0A0P5ZNF8 A0A0P6BKM0 A0A158NT58 A0A195BHJ0 E2BA17 A0A0A9XBQ8 A0A0A9XGN9 E0VBJ3 A0A0P4VJH9 A0A0K8TJE3 A0A195EZP2 A0A0P4YV70 A0A2H8TT86
A0A2H1VM50 U5ERN6 A0A336KTF1 A0A1Q3FJ03 Q17AU9 A0A1Q3FJ69 B0W3Q5 Q7PNM1 A0A1I8JVP8 A0A067REN7 A0A1J1HXI6 A0A2M4BLW0 D6WNC0 A0A084WE82 W5J8W1 A0A1Y1LT21 A0A2J7R2R2 A0A2M3Z4T4 A0A1Y1LP75 A0A1Y1LR45 A0A2J7R2Q8 A0A0L0C1H5 A0A154P3V2 A0A1I8MDL9 T1P881 B3MHR6 A0A0M4EKR8 B4H8I7 A0A0L7LGQ7 Q28YK0 B4QEP7 B4HR44 Q7K1D7 A0A3B0JPZ5 B3N9W4 A0A1W4VBL3 B4P1S4 B4LJG3 B4J7Y9 A0A1I8PJP5 K7J1E9 B4KPQ6 A0A182QSK1 A0A1B6KB51 A0A1B6E568 A0A1B6ECM9 A0A1B6H316 A0A1B6EIH7 A0A1B6CJ43 B4MIP5 A0A1B6DT76 E9HG13 A0A1B6EH14 A0A232FET5 V5G4X6 A0A0A1XE15 A0A0L7R1Y5 A0A026WQE7 A0A2S2Q2E4 A0A0N8CTS5 A0A310SIR3 A0A0P5V8Q8 A0A0P5Z5F3 A0A0P5E5C2 E9H209 A0A0C9RFK2 C4WRM7 A0A0P6DT70 A0A2S2PT36 A0A1A9ZYG6 A0A0C9R6F8 A0A088A629 A0A0P5GFG3 A0A0P4XNY6 A0A0P6HJA0 V9I6K4 J9JQF9 V9I6M3 W8BIC4 A0A2A3EIP9 A0A0P5ZNF8 A0A0P6BKM0 A0A158NT58 A0A195BHJ0 E2BA17 A0A0A9XBQ8 A0A0A9XGN9 E0VBJ3 A0A0P4VJH9 A0A0K8TJE3 A0A195EZP2 A0A0P4YV70 A0A2H8TT86
Pubmed
22118469
26354079
28756777
19121390
17510324
12364791
+ More
14747013 17210077 24845553 18362917 19820115 24438588 20920257 23761445 28004739 26108605 25315136 17994087 18057021 26227816 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20075255 21292972 28648823 25830018 24508170 30249741 24495485 21347285 20798317 25401762 20566863 27129103
14747013 17210077 24845553 18362917 19820115 24438588 20920257 23761445 28004739 26108605 25315136 17994087 18057021 26227816 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20075255 21292972 28648823 25830018 24508170 30249741 24495485 21347285 20798317 25401762 20566863 27129103
EMBL
AGBW02009953
OWR49506.1
NWSH01005933
PCG63830.1
KQ459465
KPJ00323.1
+ More
KZ150214 PZC72144.1 KQ460393 KPJ15363.1 BABH01016236 BABH01016237 BABH01016238 BABH01016239 ODYU01003316 SOQ41909.1 GANO01003661 JAB56210.1 UFQS01000243 UFQS01000465 UFQT01000243 UFQT01000465 SSX01907.1 SSX04230.1 SSX22284.1 GFDL01007434 JAV27611.1 CH477330 EAT43359.1 GFDL01007426 JAV27619.1 DS231833 EDS32202.1 AAAB01008960 EAA11941.4 KK852744 KDR17346.1 CVRI01000035 CRK92699.1 GGFJ01004925 MBW54066.1 KQ971343 EFA03239.1 ATLV01023193 ATLV01023194 KE525341 KFB48526.1 ADMH02002096 ETN59239.1 GEZM01050662 JAV75440.1 NEVH01007828 PNF35121.1 GGFM01002773 MBW23524.1 GEZM01050663 JAV75439.1 GEZM01050661 JAV75441.1 PNF35120.1 JRES01001012 KNC26140.1 KQ434809 KZC06596.1 KA644794 AFP59423.1 CH902619 EDV36903.1 KPU76500.1 KPU76501.1 KPU76502.1 CP012524 ALC42030.1 CH479223 EDW35022.1 JTDY01001256 KOB74381.1 CM000071 EAL25965.1 KRT02709.1 KRT02710.1 CM000362 CM002911 EDX06043.1 KMY92027.1 CH480816 EDW46787.1 AE013599 AY069142 BT120089 AAF59224.1 AAL39287.1 ADB04946.1 OUUW01000001 SPP75729.1 CH954177 EDV59660.1 CM000157 EDW89210.1 CH940648 EDW61531.1 KRF80040.1 KRF80041.1 KRF80042.1 CH916367 EDW02219.1 CH933808 EDW09166.1 KRG04481.1 KRG04482.1 KRG04483.1 AXCN02000446 GEBQ01031305 JAT08672.1 GEDC01004226 JAS33072.1 GEDC01001603 JAS35695.1 GECZ01027175 GECZ01023503 GECZ01000685 JAS42594.1 JAS46266.1 JAS69084.1 GECZ01032039 GECZ01028696 JAS37730.1 JAS41073.1 GEDC01023782 JAS13516.1 CH963719 EDW71984.1 GEDC01008459 JAS28839.1 GL732638 EFX69311.1 GEDC01000079 JAS37219.1 NNAY01000330 OXU29172.1 GALX01003366 JAB65100.1 GBXI01014800 GBXI01005107 GBXI01003454 JAC99491.1 JAD09185.1 JAD10838.1 KQ414667 KOC64839.1 KK107132 QOIP01000009 EZA58153.1 RLU18697.1 GGMS01002735 MBY71938.1 GDIP01099647 LRGB01003375 JAM04068.1 KZS02998.1 KQ759828 OAD62668.1 GDIP01118747 JAL84967.1 GDIP01048954 JAM54761.1 GDIP01146797 JAJ76605.1 GL732584 EFX74262.1 GBYB01011907 JAG81674.1 AK339832 BAH70547.1 GDIQ01073254 JAN21483.1 GGMR01019990 MBY32609.1 GBYB01011909 JAG81676.1 GDIQ01241695 JAK10030.1 GDIP01241226 LRGB01000725 JAI82175.1 KZS16272.1 GDIQ01018898 JAN75839.1 JR035733 JR035734 AEY56643.1 AEY56644.1 ABLF02022581 ABLF02022583 ABLF02022584 JR035725 JR035726 JR035729 JR035732 AEY56635.1 AEY56636.1 AEY56639.1 AEY56642.1 GAMC01013550 GAMC01013549 JAB93006.1 KZ288229 PBC31673.1 GDIP01041899 JAM61816.1 GDIP01020936 JAM82779.1 ADTU01025488 KQ976488 KYM83619.1 GL446664 EFN87426.1 GBHO01027356 JAG16248.1 GBHO01027354 JAG16250.1 DS235031 EEB10749.1 GDKW01002242 JAI54353.1 GBRD01000320 JAG65501.1 KQ981897 KYN33693.1 GDIP01223180 JAJ00222.1 GFXV01004997 MBW16802.1
KZ150214 PZC72144.1 KQ460393 KPJ15363.1 BABH01016236 BABH01016237 BABH01016238 BABH01016239 ODYU01003316 SOQ41909.1 GANO01003661 JAB56210.1 UFQS01000243 UFQS01000465 UFQT01000243 UFQT01000465 SSX01907.1 SSX04230.1 SSX22284.1 GFDL01007434 JAV27611.1 CH477330 EAT43359.1 GFDL01007426 JAV27619.1 DS231833 EDS32202.1 AAAB01008960 EAA11941.4 KK852744 KDR17346.1 CVRI01000035 CRK92699.1 GGFJ01004925 MBW54066.1 KQ971343 EFA03239.1 ATLV01023193 ATLV01023194 KE525341 KFB48526.1 ADMH02002096 ETN59239.1 GEZM01050662 JAV75440.1 NEVH01007828 PNF35121.1 GGFM01002773 MBW23524.1 GEZM01050663 JAV75439.1 GEZM01050661 JAV75441.1 PNF35120.1 JRES01001012 KNC26140.1 KQ434809 KZC06596.1 KA644794 AFP59423.1 CH902619 EDV36903.1 KPU76500.1 KPU76501.1 KPU76502.1 CP012524 ALC42030.1 CH479223 EDW35022.1 JTDY01001256 KOB74381.1 CM000071 EAL25965.1 KRT02709.1 KRT02710.1 CM000362 CM002911 EDX06043.1 KMY92027.1 CH480816 EDW46787.1 AE013599 AY069142 BT120089 AAF59224.1 AAL39287.1 ADB04946.1 OUUW01000001 SPP75729.1 CH954177 EDV59660.1 CM000157 EDW89210.1 CH940648 EDW61531.1 KRF80040.1 KRF80041.1 KRF80042.1 CH916367 EDW02219.1 CH933808 EDW09166.1 KRG04481.1 KRG04482.1 KRG04483.1 AXCN02000446 GEBQ01031305 JAT08672.1 GEDC01004226 JAS33072.1 GEDC01001603 JAS35695.1 GECZ01027175 GECZ01023503 GECZ01000685 JAS42594.1 JAS46266.1 JAS69084.1 GECZ01032039 GECZ01028696 JAS37730.1 JAS41073.1 GEDC01023782 JAS13516.1 CH963719 EDW71984.1 GEDC01008459 JAS28839.1 GL732638 EFX69311.1 GEDC01000079 JAS37219.1 NNAY01000330 OXU29172.1 GALX01003366 JAB65100.1 GBXI01014800 GBXI01005107 GBXI01003454 JAC99491.1 JAD09185.1 JAD10838.1 KQ414667 KOC64839.1 KK107132 QOIP01000009 EZA58153.1 RLU18697.1 GGMS01002735 MBY71938.1 GDIP01099647 LRGB01003375 JAM04068.1 KZS02998.1 KQ759828 OAD62668.1 GDIP01118747 JAL84967.1 GDIP01048954 JAM54761.1 GDIP01146797 JAJ76605.1 GL732584 EFX74262.1 GBYB01011907 JAG81674.1 AK339832 BAH70547.1 GDIQ01073254 JAN21483.1 GGMR01019990 MBY32609.1 GBYB01011909 JAG81676.1 GDIQ01241695 JAK10030.1 GDIP01241226 LRGB01000725 JAI82175.1 KZS16272.1 GDIQ01018898 JAN75839.1 JR035733 JR035734 AEY56643.1 AEY56644.1 ABLF02022581 ABLF02022583 ABLF02022584 JR035725 JR035726 JR035729 JR035732 AEY56635.1 AEY56636.1 AEY56639.1 AEY56642.1 GAMC01013550 GAMC01013549 JAB93006.1 KZ288229 PBC31673.1 GDIP01041899 JAM61816.1 GDIP01020936 JAM82779.1 ADTU01025488 KQ976488 KYM83619.1 GL446664 EFN87426.1 GBHO01027356 JAG16248.1 GBHO01027354 JAG16250.1 DS235031 EEB10749.1 GDKW01002242 JAI54353.1 GBRD01000320 JAG65501.1 KQ981897 KYN33693.1 GDIP01223180 JAJ00222.1 GFXV01004997 MBW16802.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000005204
UP000008820
+ More
UP000002320 UP000007062 UP000076407 UP000027135 UP000183832 UP000007266 UP000030765 UP000000673 UP000235965 UP000037069 UP000076502 UP000095301 UP000007801 UP000092553 UP000008744 UP000037510 UP000001819 UP000000304 UP000001292 UP000000803 UP000268350 UP000008711 UP000192221 UP000002282 UP000008792 UP000001070 UP000095300 UP000002358 UP000009192 UP000075886 UP000007798 UP000000305 UP000215335 UP000053825 UP000053097 UP000279307 UP000076858 UP000092445 UP000005203 UP000007819 UP000242457 UP000005205 UP000078540 UP000008237 UP000009046 UP000078541
UP000002320 UP000007062 UP000076407 UP000027135 UP000183832 UP000007266 UP000030765 UP000000673 UP000235965 UP000037069 UP000076502 UP000095301 UP000007801 UP000092553 UP000008744 UP000037510 UP000001819 UP000000304 UP000001292 UP000000803 UP000268350 UP000008711 UP000192221 UP000002282 UP000008792 UP000001070 UP000095300 UP000002358 UP000009192 UP000075886 UP000007798 UP000000305 UP000215335 UP000053825 UP000053097 UP000279307 UP000076858 UP000092445 UP000005203 UP000007819 UP000242457 UP000005205 UP000078540 UP000008237 UP000009046 UP000078541
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A212F6Y2
A0A2A4IXF8
A0A194Q455
A0A2W1BEJ5
A0A0N1PIB1
H9JRD7
+ More
A0A2H1VM50 U5ERN6 A0A336KTF1 A0A1Q3FJ03 Q17AU9 A0A1Q3FJ69 B0W3Q5 Q7PNM1 A0A1I8JVP8 A0A067REN7 A0A1J1HXI6 A0A2M4BLW0 D6WNC0 A0A084WE82 W5J8W1 A0A1Y1LT21 A0A2J7R2R2 A0A2M3Z4T4 A0A1Y1LP75 A0A1Y1LR45 A0A2J7R2Q8 A0A0L0C1H5 A0A154P3V2 A0A1I8MDL9 T1P881 B3MHR6 A0A0M4EKR8 B4H8I7 A0A0L7LGQ7 Q28YK0 B4QEP7 B4HR44 Q7K1D7 A0A3B0JPZ5 B3N9W4 A0A1W4VBL3 B4P1S4 B4LJG3 B4J7Y9 A0A1I8PJP5 K7J1E9 B4KPQ6 A0A182QSK1 A0A1B6KB51 A0A1B6E568 A0A1B6ECM9 A0A1B6H316 A0A1B6EIH7 A0A1B6CJ43 B4MIP5 A0A1B6DT76 E9HG13 A0A1B6EH14 A0A232FET5 V5G4X6 A0A0A1XE15 A0A0L7R1Y5 A0A026WQE7 A0A2S2Q2E4 A0A0N8CTS5 A0A310SIR3 A0A0P5V8Q8 A0A0P5Z5F3 A0A0P5E5C2 E9H209 A0A0C9RFK2 C4WRM7 A0A0P6DT70 A0A2S2PT36 A0A1A9ZYG6 A0A0C9R6F8 A0A088A629 A0A0P5GFG3 A0A0P4XNY6 A0A0P6HJA0 V9I6K4 J9JQF9 V9I6M3 W8BIC4 A0A2A3EIP9 A0A0P5ZNF8 A0A0P6BKM0 A0A158NT58 A0A195BHJ0 E2BA17 A0A0A9XBQ8 A0A0A9XGN9 E0VBJ3 A0A0P4VJH9 A0A0K8TJE3 A0A195EZP2 A0A0P4YV70 A0A2H8TT86
A0A2H1VM50 U5ERN6 A0A336KTF1 A0A1Q3FJ03 Q17AU9 A0A1Q3FJ69 B0W3Q5 Q7PNM1 A0A1I8JVP8 A0A067REN7 A0A1J1HXI6 A0A2M4BLW0 D6WNC0 A0A084WE82 W5J8W1 A0A1Y1LT21 A0A2J7R2R2 A0A2M3Z4T4 A0A1Y1LP75 A0A1Y1LR45 A0A2J7R2Q8 A0A0L0C1H5 A0A154P3V2 A0A1I8MDL9 T1P881 B3MHR6 A0A0M4EKR8 B4H8I7 A0A0L7LGQ7 Q28YK0 B4QEP7 B4HR44 Q7K1D7 A0A3B0JPZ5 B3N9W4 A0A1W4VBL3 B4P1S4 B4LJG3 B4J7Y9 A0A1I8PJP5 K7J1E9 B4KPQ6 A0A182QSK1 A0A1B6KB51 A0A1B6E568 A0A1B6ECM9 A0A1B6H316 A0A1B6EIH7 A0A1B6CJ43 B4MIP5 A0A1B6DT76 E9HG13 A0A1B6EH14 A0A232FET5 V5G4X6 A0A0A1XE15 A0A0L7R1Y5 A0A026WQE7 A0A2S2Q2E4 A0A0N8CTS5 A0A310SIR3 A0A0P5V8Q8 A0A0P5Z5F3 A0A0P5E5C2 E9H209 A0A0C9RFK2 C4WRM7 A0A0P6DT70 A0A2S2PT36 A0A1A9ZYG6 A0A0C9R6F8 A0A088A629 A0A0P5GFG3 A0A0P4XNY6 A0A0P6HJA0 V9I6K4 J9JQF9 V9I6M3 W8BIC4 A0A2A3EIP9 A0A0P5ZNF8 A0A0P6BKM0 A0A158NT58 A0A195BHJ0 E2BA17 A0A0A9XBQ8 A0A0A9XGN9 E0VBJ3 A0A0P4VJH9 A0A0K8TJE3 A0A195EZP2 A0A0P4YV70 A0A2H8TT86
Ontologies
GO
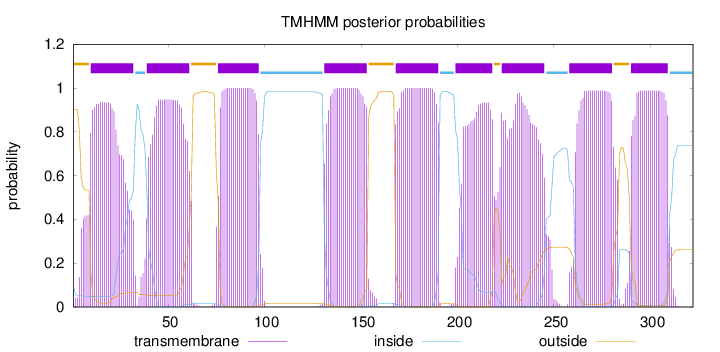
Topology
Length:
322
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
188.19601
Exp number, first 60 AAs:
39.80244
Total prob of N-in:
0.09665
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 38
TMhelix
39 - 61
outside
62 - 75
TMhelix
76 - 97
inside
98 - 130
TMhelix
131 - 153
outside
154 - 167
TMhelix
168 - 190
inside
191 - 198
TMhelix
199 - 218
outside
219 - 222
TMhelix
223 - 245
inside
246 - 257
TMhelix
258 - 280
outside
281 - 289
TMhelix
290 - 309
inside
310 - 322
Population Genetic Test Statistics
Pi
262.871478
Theta
18.816664
Tajima's D
1.090841
CLR
0.380052
CSRT
0.689665516724164
Interpretation
Uncertain
