Gene
KWMTBOMO02405
Annotation
PREDICTED:_uncharacterized_protein_K02A2.6-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.404
Sequence
CDS
ATGGAACACGCCCGACCACCGTCAGAATTGAGTCTGGAAGGCAACTCCGTCAGCCGGGCTGACGCATGGAAGCGTTGGAGGAAACAATTTCATGTATTTTTGAAAGCATCAGGGGTGCATAAAGAAGAAGGCAGTGTGCAAGCCAGCTTGCTCATCAATTTAATTGGAACGGAAGGTTTCGATGTGTACGAGACGTTTAAGTTTTCAAATGACACTGAAAAAGATGACGTAACGGTTTTGTTAAAAAAATTCGACGAATACTTCGGCGTCAAACCAAATGTTACGTTGGCGAGATATAATTTCTACATGAGAAACCAGGAAGGCGGCGAGTCCATAGATCAGTATGTGACGGCTTTGAAACTGCTGAGCAAAACCTGTGAATTTAAAACCTTAGAAGACGAATTCATTCGCGATCGGATCGTATGTGGCCTAAAAAATACGATAGTTCGCGATCGTTTACTGAGAACGGACGACCTGACGATGGACAAAGCTATTAAAATATGCCAAGTCGACGAGGTATCGGCGGACAGCAGTCGCGTATTGCAGGAGTCGCGCGGAGCGAGCGCGGAGCCGGTGCGCGTGGACGCTGTGGGCGCGCGGGGTGGTGCTGGGCCACGTGCGGCGCGGGGCCGAGCCGGCGGCGCGCGAGCAGCTCGCGGCGGCCGCGGCGGTCACCGGCTGCGTGCGGTCCCGGATGGCGCGGGGCGCCCGCCGCCTCTTTCCAGTTCATCTGCGTCCAGTACTTGTCTCAGGTGCGGAGGTCCGTGTGTTAGTTGGGCTCACTGTCCTGCGGTCTCAGTGCAGTGTTATGTGTGTCAAAATTATGGTCATTTTGCAAGAATGTGTCAAGTACATAAAGGTGTAAAAAAGTTATATGATATCGGTGTTCTAGAGGACGAAAATAAATACGAGCCGGAACTGGACGATGACTGGGAGTCGTTTTATATATCTACTTTAATTGATGGTTCCAAAATCGACTCGTTGTCGCAGGATTGGTTTGAGGTCCTCCGAGGAGAATGGGGCTCGGAAAATTTTAAACTAGACTCGGGTGCAGATATCAATGTCTTATCTTTTAAGCGATTTGTGCAGTTAGGCAATAACCCTAATAACATTGTTGTAAATCATAAGGTTAAACTGCAATCTTACAGTGGCGATTTCATTCCTATTAAGGGAATTTGTAATATTAAATGGTGGTATAAAAATGTTCAATATGATCTTAAGTTTGCGATAGCTGATATTGATTGTCAAAGTGTACTGGGACTGCAGGCGTGCTTATTATTAGGGTTAATAAAAAGAATTCACGATATAAATTTCTCTAAAAATAGTGATTTGTTCACTGGGTTAGGTTGTTTACCGGGGGAATATCATATAACGGTAGATAGGGATGTGACGCCGGTGGTGTGCGCTCCGCGGAAAGTACCGTTAAGCCTTCGTGATAACTTGAAGGACGAACTCGATAGGATGACAGAGTTAGGTGTGATTAGGAAGGTGACTCATCCGACACCTTGGGTCAACTCGTTAGTAGTAGTGGCCAAGACAAATGGTAAAATGCGCATATGCCTAGACCCGAGACCTCTCAATAAGGCGATTCAACGTGCTCATTTTCAACTACCCACCATAAATGAGTTGGCTACCAAATTGAATGGGGCTAAATATTTTTCGGTATTGGACGCCAATTCAGGATTTTGGTCCGTAAAGCTAGATAAGGAGAGTGCCGATTTATGCACATTTATTACGCCGTTTGGGCGATATCAATATCTAAGATTACCGTTCGGACTGAACTGCGCACCGGAGGTATTCCACGCTAAATTAAAACAATTGTTAGAAGGTTTAGACGGTGTTGAGTCGTTTATTGATGATATTATTGTCTGGGGGTCCACTAGGCGAGAACATGATGTAAGACTAAATGCGTTATTTCAAAAAGCGCGAGATATTAATTTAAAGTTTAACAAAGATAAGTGTCGCATTTGTGTCGATGAGGTGACATATTTGGGACATATTTTTAATAAAGACGGAATGAAAGTAGATACGGAAAAAGTGCGGGCTGTAATCAATATGCCGGAACCTACGGATTGTAAAAGTTTAGAAAGGTTCTTGGGTGCAATTAATTATTTATCAAAGTTTATACCTAACTACTCGCAACACATATTTCCATTGACTAGATTATTGAAAAAAGATGTCGAGTGGTGTTGGGAAATAGGTCACAAAAACGCCTTTGATAAATTAAAGAAACTGATATGCGAGGCTCCGGTATTATCGTTGTTTGACGTAGGCAGTGAGGTATTGTTGTCGGTGGACGCGAGCAGCGTCGCACTTGGCGCGGTGCTGATGCAGCGCGGCCGCCCGGTCGAGTACGCGTCGCGCACACTCACCGACACACAACAGAGATACGCTCAGATTGAAAAAGAAATGTTAGCTATTGTCTTTGCTTGTGAAAAATTTCACCAGTACATTTATGGAAAAAGGTATATAGTCATAGAAACTGACCATAAACCTCTTGAAAGTATTTTTAAAAAGCCGCTCAATTCAGTCCCAGCACGCTTGCAGCGTATGATGTTAAAATTACAAGGGTACGATCTGAAAATTACTTATAAACCGGGGAAGTATATGTATATACCCGACACGTTATCACGTGCGCCTCTTCCGGATTTATATGACGATAAAATTAGTAAGGCAGTATTGAACCAATTAAAAATGGTAATTAACAATGTACCGATGTCTCAAAGCAAACTGACATTAGTTAAGAAAGAAACTAGTAAAGATGAGTATTTGAAACGTTTGTTAGTTTACATTAATGAAGGTTGGCCCGTTCATAAATATAATGTTGATAGTGATTTAAAGTATTTTTGGTCCATGAGAGACGAGCTGTACTCTGTTGACGGAGTAATTTTTAAAGATAAACAGCAACAGCATAAAACAATAATAGAAAAACAATTAAAAAGTAAAATGTATTATGACCAACATTGTAAAGAGCAGCCGGAGCTAAGTGTCGGCGATGATGTAATTATGTTAGAACATAATAATGAACGAATGCGCGGCACTGTCGTAGGCAAGGCATCTGCACCGCGTTCATACATTGTAAAAAACAAATTAGGTATATATCGACGTAATCGCCGACATTTAATTAAAAATATGCCTGATGAGAAACCTAATGTTGTGGAAAAGCTGAAGAATGATCGGCGCGGCACAATAGTGATCGTATCAGACGAGTCAAGTGGGGGAGAATATGAGATGAGTGTGGATGAGGATGGTTCAGTGTACGAACCGAGTACTTTGTCATCACCTTCATCACCTCATATTTATCCGCCTACTACTCGTAGTAGAACTAAGCAGAATGCAATTAATAATAATTAA
Protein
MEHARPPSELSLEGNSVSRADAWKRWRKQFHVFLKASGVHKEEGSVQASLLINLIGTEGFDVYETFKFSNDTEKDDVTVLLKKFDEYFGVKPNVTLARYNFYMRNQEGGESIDQYVTALKLLSKTCEFKTLEDEFIRDRIVCGLKNTIVRDRLLRTDDLTMDKAIKICQVDEVSADSSRVLQESRGASAEPVRVDAVGARGGAGPRAARGRAGGARAARGGRGGHRLRAVPDGAGRPPPLSSSSASSTCLRCGGPCVSWAHCPAVSVQCYVCQNYGHFARMCQVHKGVKKLYDIGVLEDENKYEPELDDDWESFYISTLIDGSKIDSLSQDWFEVLRGEWGSENFKLDSGADINVLSFKRFVQLGNNPNNIVVNHKVKLQSYSGDFIPIKGICNIKWWYKNVQYDLKFAIADIDCQSVLGLQACLLLGLIKRIHDINFSKNSDLFTGLGCLPGEYHITVDRDVTPVVCAPRKVPLSLRDNLKDELDRMTELGVIRKVTHPTPWVNSLVVVAKTNGKMRICLDPRPLNKAIQRAHFQLPTINELATKLNGAKYFSVLDANSGFWSVKLDKESADLCTFITPFGRYQYLRLPFGLNCAPEVFHAKLKQLLEGLDGVESFIDDIIVWGSTRREHDVRLNALFQKARDINLKFNKDKCRICVDEVTYLGHIFNKDGMKVDTEKVRAVINMPEPTDCKSLERFLGAINYLSKFIPNYSQHIFPLTRLLKKDVEWCWEIGHKNAFDKLKKLICEAPVLSLFDVGSEVLLSVDASSVALGAVLMQRGRPVEYASRTLTDTQQRYAQIEKEMLAIVFACEKFHQYIYGKRYIVIETDHKPLESIFKKPLNSVPARLQRMMLKLQGYDLKITYKPGKYMYIPDTLSRAPLPDLYDDKISKAVLNQLKMVINNVPMSQSKLTLVKKETSKDEYLKRLLVYINEGWPVHKYNVDSDLKYFWSMRDELYSVDGVIFKDKQQQHKTIIEKQLKSKMYYDQHCKEQPELSVGDDVIMLEHNNERMRGTVVGKASAPRSYIVKNKLGIYRRNRRHLIKNMPDEKPNVVEKLKNDRRGTIVIVSDESSGGEYEMSVDEDGSVYEPSTLSSPSSPHIYPPTTRSRTKQNAINNN
Summary
Uniprot
A0A3P8Q6C6
A0A3B3HLX7
A0A2G8KFU3
A0A3B1JKR1
A0A3P8S414
A0A2G8KHQ8
+ More
A0A2S2N8I3 A0A2G8LMB1 A0A023EY30 A0A3P9HT22 A0A1A8UGA0 A0A3B3C9X5 X1WTY2 A0A0A9Z4Y4 A0A1Y1MZH6 A0A146LC45 A0A1Y1L7V1 A0A3P9LZF9 A0A3P9K9H2 A0A069DZS8 A0A3P8RHF8 A0A069DY08 A0A3P8Q2Q2 A0A1A8DQM9 A0A3P8RKT7 A0A1A8S1T9 A0A3P9AYU2 A0A3P9HXP1 A0A1A8L2U7 J9L288 A0A146TAZ6 A0A3P8R8M6 A0A3B3I1S4 A0A1S3DJN8 A0A0P6BAV5 A0A0P5A948 A0A0P5V4F7 A0A147BKH7 A0A0P4XRP4 A0A0P5XDK7 A0A3Q0JJE3 A0A0P5VCW1 A0A0P5DHJ0 A0A1S3JEC7 A0A1Y1MCJ7 A0A3P8QAD1 A0A267GLM4 I3KYE2 A0A146SN99 A0A1A8N8J3 A0A147BIE4 A0A3P9L920 A0A267GXW3 A0A1Y1M671 A0A1S3HHB2 A0A267DET5 A0A1Y1KVT8 A0A147BJX4 A0A147BN28 W4YP04 A0A1Y1NDC5 A0A0A9W3X9 A0A075EIL5 A0A1S3DSZ0 A0A146L0H7 W4Y1E4 W4YGV7 A0A1Y1LVD1 A0A1S3DKC8 A0A2S2NI46 J9LYD2 A0A0A9VTL5 A0A3P9JKX9 A0A3P9HMN7 A0A1Y1JV53
A0A2S2N8I3 A0A2G8LMB1 A0A023EY30 A0A3P9HT22 A0A1A8UGA0 A0A3B3C9X5 X1WTY2 A0A0A9Z4Y4 A0A1Y1MZH6 A0A146LC45 A0A1Y1L7V1 A0A3P9LZF9 A0A3P9K9H2 A0A069DZS8 A0A3P8RHF8 A0A069DY08 A0A3P8Q2Q2 A0A1A8DQM9 A0A3P8RKT7 A0A1A8S1T9 A0A3P9AYU2 A0A3P9HXP1 A0A1A8L2U7 J9L288 A0A146TAZ6 A0A3P8R8M6 A0A3B3I1S4 A0A1S3DJN8 A0A0P6BAV5 A0A0P5A948 A0A0P5V4F7 A0A147BKH7 A0A0P4XRP4 A0A0P5XDK7 A0A3Q0JJE3 A0A0P5VCW1 A0A0P5DHJ0 A0A1S3JEC7 A0A1Y1MCJ7 A0A3P8QAD1 A0A267GLM4 I3KYE2 A0A146SN99 A0A1A8N8J3 A0A147BIE4 A0A3P9L920 A0A267GXW3 A0A1Y1M671 A0A1S3HHB2 A0A267DET5 A0A1Y1KVT8 A0A147BJX4 A0A147BN28 W4YP04 A0A1Y1NDC5 A0A0A9W3X9 A0A075EIL5 A0A1S3DSZ0 A0A146L0H7 W4Y1E4 W4YGV7 A0A1Y1LVD1 A0A1S3DKC8 A0A2S2NI46 J9LYD2 A0A0A9VTL5 A0A3P9JKX9 A0A3P9HMN7 A0A1Y1JV53
Pubmed
EMBL
MRZV01000618
PIK46835.1
MRZV01000576
PIK47528.1
GGMR01000868
MBY13487.1
+ More
MRZV01000033 PIK61397.1 GBBI01004745 JAC13967.1 HADY01023773 HAEJ01006887 SBS47344.1 ABLF02013930 ABLF02013934 GBHO01004090 JAG39514.1 GEZM01016880 JAV91071.1 GDHC01013957 JAQ04672.1 GEZM01066276 JAV68015.1 GBGD01000115 JAC88774.1 GBGD01000119 JAC88770.1 HADZ01002490 HAEA01007842 SBQ36322.1 HAEH01020882 SBS11479.1 HAEF01001580 SBR38962.1 ABLF02022355 ABLF02022359 ABLF02022361 ABLF02022365 ABLF02041467 GCES01096219 JAQ90103.1 GDIP01019161 JAM84554.1 GDIP01203362 JAJ20040.1 GDIP01199810 GDIP01181260 GDIP01124130 GDIP01104931 GDIP01053137 GDIP01051184 JAL98783.1 GEGO01004116 JAR91288.1 GDIP01239061 JAI84340.1 GDIP01073678 JAM30037.1 GDIP01102077 JAM01638.1 GDIP01156216 JAJ67186.1 GEZM01035137 JAV83383.1 NIVC01000291 PAA86189.1 AERX01070080 GCES01104253 JAQ82069.1 HAEH01000697 SBR65395.1 GEGO01004841 JAR90563.1 NIVC01000102 PAA90881.1 GEZM01041944 JAV80090.1 NIVC01004589 PAA47069.1 GEZM01074317 JAV64601.1 GEGO01004317 JAR91087.1 GEGO01003204 JAR92200.1 AAGJ04126635 GEZM01011167 JAV93617.1 GBHO01042421 GBHO01026477 JAG01183.1 JAG17127.1 KF319019 AIE48224.1 GDHC01017753 JAQ00876.1 AAGJ04066141 AAGJ04164197 GEZM01051395 JAV74977.1 GGMR01004179 MBY16798.1 ABLF02041541 GBHO01044535 JAF99068.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1
MRZV01000033 PIK61397.1 GBBI01004745 JAC13967.1 HADY01023773 HAEJ01006887 SBS47344.1 ABLF02013930 ABLF02013934 GBHO01004090 JAG39514.1 GEZM01016880 JAV91071.1 GDHC01013957 JAQ04672.1 GEZM01066276 JAV68015.1 GBGD01000115 JAC88774.1 GBGD01000119 JAC88770.1 HADZ01002490 HAEA01007842 SBQ36322.1 HAEH01020882 SBS11479.1 HAEF01001580 SBR38962.1 ABLF02022355 ABLF02022359 ABLF02022361 ABLF02022365 ABLF02041467 GCES01096219 JAQ90103.1 GDIP01019161 JAM84554.1 GDIP01203362 JAJ20040.1 GDIP01199810 GDIP01181260 GDIP01124130 GDIP01104931 GDIP01053137 GDIP01051184 JAL98783.1 GEGO01004116 JAR91288.1 GDIP01239061 JAI84340.1 GDIP01073678 JAM30037.1 GDIP01102077 JAM01638.1 GDIP01156216 JAJ67186.1 GEZM01035137 JAV83383.1 NIVC01000291 PAA86189.1 AERX01070080 GCES01104253 JAQ82069.1 HAEH01000697 SBR65395.1 GEGO01004841 JAR90563.1 NIVC01000102 PAA90881.1 GEZM01041944 JAV80090.1 NIVC01004589 PAA47069.1 GEZM01074317 JAV64601.1 GEGO01004317 JAR91087.1 GEGO01003204 JAR92200.1 AAGJ04126635 GEZM01011167 JAV93617.1 GBHO01042421 GBHO01026477 JAG01183.1 JAG17127.1 KF319019 AIE48224.1 GDHC01017753 JAQ00876.1 AAGJ04066141 AAGJ04164197 GEZM01051395 JAV74977.1 GGMR01004179 MBY16798.1 ABLF02041541 GBHO01044535 JAF99068.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1
Proteomes
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR005162 Retrotrans_gag_dom
IPR036875 Znf_CCHC_sf
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR005162 Retrotrans_gag_dom
IPR036875 Znf_CCHC_sf
Gene 3D
ProteinModelPortal
A0A3P8Q6C6
A0A3B3HLX7
A0A2G8KFU3
A0A3B1JKR1
A0A3P8S414
A0A2G8KHQ8
+ More
A0A2S2N8I3 A0A2G8LMB1 A0A023EY30 A0A3P9HT22 A0A1A8UGA0 A0A3B3C9X5 X1WTY2 A0A0A9Z4Y4 A0A1Y1MZH6 A0A146LC45 A0A1Y1L7V1 A0A3P9LZF9 A0A3P9K9H2 A0A069DZS8 A0A3P8RHF8 A0A069DY08 A0A3P8Q2Q2 A0A1A8DQM9 A0A3P8RKT7 A0A1A8S1T9 A0A3P9AYU2 A0A3P9HXP1 A0A1A8L2U7 J9L288 A0A146TAZ6 A0A3P8R8M6 A0A3B3I1S4 A0A1S3DJN8 A0A0P6BAV5 A0A0P5A948 A0A0P5V4F7 A0A147BKH7 A0A0P4XRP4 A0A0P5XDK7 A0A3Q0JJE3 A0A0P5VCW1 A0A0P5DHJ0 A0A1S3JEC7 A0A1Y1MCJ7 A0A3P8QAD1 A0A267GLM4 I3KYE2 A0A146SN99 A0A1A8N8J3 A0A147BIE4 A0A3P9L920 A0A267GXW3 A0A1Y1M671 A0A1S3HHB2 A0A267DET5 A0A1Y1KVT8 A0A147BJX4 A0A147BN28 W4YP04 A0A1Y1NDC5 A0A0A9W3X9 A0A075EIL5 A0A1S3DSZ0 A0A146L0H7 W4Y1E4 W4YGV7 A0A1Y1LVD1 A0A1S3DKC8 A0A2S2NI46 J9LYD2 A0A0A9VTL5 A0A3P9JKX9 A0A3P9HMN7 A0A1Y1JV53
A0A2S2N8I3 A0A2G8LMB1 A0A023EY30 A0A3P9HT22 A0A1A8UGA0 A0A3B3C9X5 X1WTY2 A0A0A9Z4Y4 A0A1Y1MZH6 A0A146LC45 A0A1Y1L7V1 A0A3P9LZF9 A0A3P9K9H2 A0A069DZS8 A0A3P8RHF8 A0A069DY08 A0A3P8Q2Q2 A0A1A8DQM9 A0A3P8RKT7 A0A1A8S1T9 A0A3P9AYU2 A0A3P9HXP1 A0A1A8L2U7 J9L288 A0A146TAZ6 A0A3P8R8M6 A0A3B3I1S4 A0A1S3DJN8 A0A0P6BAV5 A0A0P5A948 A0A0P5V4F7 A0A147BKH7 A0A0P4XRP4 A0A0P5XDK7 A0A3Q0JJE3 A0A0P5VCW1 A0A0P5DHJ0 A0A1S3JEC7 A0A1Y1MCJ7 A0A3P8QAD1 A0A267GLM4 I3KYE2 A0A146SN99 A0A1A8N8J3 A0A147BIE4 A0A3P9L920 A0A267GXW3 A0A1Y1M671 A0A1S3HHB2 A0A267DET5 A0A1Y1KVT8 A0A147BJX4 A0A147BN28 W4YP04 A0A1Y1NDC5 A0A0A9W3X9 A0A075EIL5 A0A1S3DSZ0 A0A146L0H7 W4Y1E4 W4YGV7 A0A1Y1LVD1 A0A1S3DKC8 A0A2S2NI46 J9LYD2 A0A0A9VTL5 A0A3P9JKX9 A0A3P9HMN7 A0A1Y1JV53
PDB
4OL8
E-value=5.51694e-53,
Score=529
Ontologies
KEGG
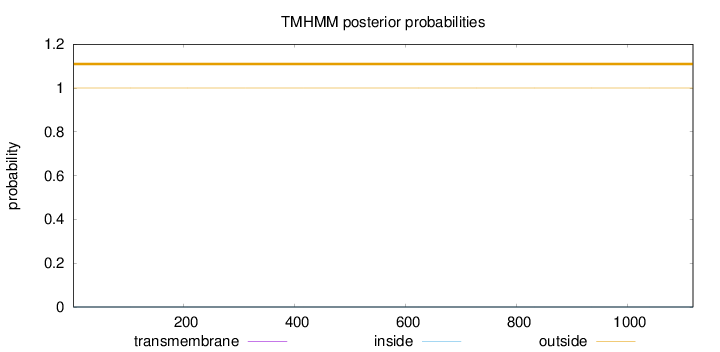
Topology
Length:
1117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00111
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00002
outside
1 - 1117
Population Genetic Test Statistics
Pi
1.770224
Theta
10.87095
Tajima's D
-1.732444
CLR
142.343148
CSRT
0.031948402579871
Interpretation
Uncertain
