Gene
KWMTBOMO02402
Pre Gene Modal
BGIBMGA013486
Annotation
PREDICTED:_NEDD8-conjugating_enzyme_UBE2F-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.207
Sequence
CDS
ATGATAACCCTTAATAGGAAACTTAAAAAGGAGCATTCTCCTCCAACAATAGGCCTGACCACGGAGCCAGTTAAAAGGATATCAATACGAGATAAATTGTTAGTAAAAGAAGTACAGGAGATGGAAGAGAACCTTCCAGCAACTTGTAGCGTGAAGTTCGAAGACCCAAATCTGCTTAGTGAATTCATATTAACAGTAGCTCCTGATGAGGGGTATTGGAAAGGTGGTAAATTTAAGTTTAGTGTCATTATAACTGAAGAATATAATATGGCTCCACCAAGAGTGAAGTGTTTAACAAAATTATGGCATCCTAATATTAATGTAGATGGAGACATCTGTTTGTCATTACTGAGACAGACTTCTATTGATGAGCATGGCTGGGCACCTACTAGGAGGCTCAAAGATGTTGTATGGGGCTTGAACTCACTTTTTACGGATCTGCTTAACTTTGATGACCCGTTAAACATAGAAGCAGCTGAAATGTACAACAAGGACAAAGTTCAATTTCAGGTTAAGGTACAGGAATATATTTCAAAGGGGAAAAGATGA
Protein
MITLNRKLKKEHSPPTIGLTTEPVKRISIRDKLLVKEVQEMEENLPATCSVKFEDPNLLSEFILTVAPDEGYWKGGKFKFSVIITEEYNMAPPRVKCLTKLWHPNINVDGDICLSLLRQTSIDEHGWAPTRRLKDVVWGLNSLFTDLLNFDDPLNIEAAEMYNKDKVQFQVKVQEYISKGKR
Summary
Similarity
Belongs to the ubiquitin-conjugating enzyme family.
Uniprot
H9JVC3
A0A2H1VI57
A0A2A4JL87
A0A2W1BDL2
A0A1E1WFH9
A0A194Q460
+ More
A0A212ELL7 A0A0N0PCZ1 A0A2P8XMW8 A0A2J7QC45 A0A1B6D9C5 Q16ND7 A0A1B6DYY7 A0A1W7R7U1 A0A2M4AYF4 A0A182K9U4 A0A084WUS3 R4UN35 A0A1Q3G2P1 A0A1Q3G2M3 A0A182N2L3 A0A1Q3G2B6 A0A1B6F4U1 A0A2M4C0Z9 A0A182IKF2 W5JGP6 A0A182QWZ2 A0A182Y219 A0A067RKI0 A0A182RYR4 A0A182MS10 A0A182W9D9 A0A2M3ZJT1 A0A182P1S5 A0A182FKM7 A0A182WZY1 A0A182HPU7 A0A182VDZ4 A0T4G7 A0A1Q3G1V9 A0A1Q3G2L0 A0A336MI34 A0A1Q3G1W8 A0A1Q3G1P4 K7IR81 A0A2A3EIP1 A0A088A625 U5ESE0 A0A0L7R253 A0A0P6CYD8 A0A0P5QNP6 A0A0P5WTM9 A0A0P5WZI5 T1IP91 A0A0P5MJG0 A0A0P5DH13 A0A0N8AEP1 A0A310SSR5 E9GAU8 A0A1B0D5J1 A0A023FAT8 A0A224XZC7 A0A069DNV1 A0A0V0GBZ4 A0A0P4VYN8 R4G407 K1RU74 E2AYP0 F4WYU7 A0A232EVY7 A0A195F9B2 A0A151JBN7 A0A195CN95 A0A158NC59 A0A026WSW3 A0A0A9W0Y3 A0A1S3IHD5 A0A0P5N8D4 A0A154P5H3 E9IM20 A0A087TNN0 A0A0P5JK99 A0A0P5V2H9 A0A0N8BBM3 A0A182UCT1 E2B5C3 A0A1L8DSV9 A0A0P5ANV1 E0VAH4 A0A1B0CNM0 A0A210Q6I6 A0A0P6CF42 A0A1Z5L9W8 A0A2R5LA70 A0A1E1XVB2 G3TCR7 A0A0L8GC71 A0A0M9AC34 A0A0C9SB84 G3I238
A0A212ELL7 A0A0N0PCZ1 A0A2P8XMW8 A0A2J7QC45 A0A1B6D9C5 Q16ND7 A0A1B6DYY7 A0A1W7R7U1 A0A2M4AYF4 A0A182K9U4 A0A084WUS3 R4UN35 A0A1Q3G2P1 A0A1Q3G2M3 A0A182N2L3 A0A1Q3G2B6 A0A1B6F4U1 A0A2M4C0Z9 A0A182IKF2 W5JGP6 A0A182QWZ2 A0A182Y219 A0A067RKI0 A0A182RYR4 A0A182MS10 A0A182W9D9 A0A2M3ZJT1 A0A182P1S5 A0A182FKM7 A0A182WZY1 A0A182HPU7 A0A182VDZ4 A0T4G7 A0A1Q3G1V9 A0A1Q3G2L0 A0A336MI34 A0A1Q3G1W8 A0A1Q3G1P4 K7IR81 A0A2A3EIP1 A0A088A625 U5ESE0 A0A0L7R253 A0A0P6CYD8 A0A0P5QNP6 A0A0P5WTM9 A0A0P5WZI5 T1IP91 A0A0P5MJG0 A0A0P5DH13 A0A0N8AEP1 A0A310SSR5 E9GAU8 A0A1B0D5J1 A0A023FAT8 A0A224XZC7 A0A069DNV1 A0A0V0GBZ4 A0A0P4VYN8 R4G407 K1RU74 E2AYP0 F4WYU7 A0A232EVY7 A0A195F9B2 A0A151JBN7 A0A195CN95 A0A158NC59 A0A026WSW3 A0A0A9W0Y3 A0A1S3IHD5 A0A0P5N8D4 A0A154P5H3 E9IM20 A0A087TNN0 A0A0P5JK99 A0A0P5V2H9 A0A0N8BBM3 A0A182UCT1 E2B5C3 A0A1L8DSV9 A0A0P5ANV1 E0VAH4 A0A1B0CNM0 A0A210Q6I6 A0A0P6CF42 A0A1Z5L9W8 A0A2R5LA70 A0A1E1XVB2 G3TCR7 A0A0L8GC71 A0A0M9AC34 A0A0C9SB84 G3I238
Pubmed
19121390
28756777
26354079
22118469
29403074
17510324
+ More
24438588 20920257 23761445 25244985 24845553 12364791 14747013 17210077 20075255 21292972 25474469 26334808 27129103 22992520 20798317 21719571 28648823 21347285 24508170 30249741 25401762 26823975 21282665 20566863 28812685 28528879 29209593 26131772 21804562
24438588 20920257 23761445 25244985 24845553 12364791 14747013 17210077 20075255 21292972 25474469 26334808 27129103 22992520 20798317 21719571 28648823 21347285 24508170 30249741 25401762 26823975 21282665 20566863 28812685 28528879 29209593 26131772 21804562
EMBL
BABH01036148
ODYU01002690
SOQ40519.1
NWSH01001115
PCG72536.1
KZ150114
+ More
PZC73292.1 GDQN01005433 JAT85621.1 KQ459465 KPJ00328.1 AGBW02014024 OWR42382.1 KQ460393 KPJ15357.1 PYGN01001693 PSN33336.1 NEVH01016292 PNF26151.1 GEDC01015014 GEDC01005778 JAS22284.1 JAS31520.1 CH477828 EAT35862.1 GEDC01006396 JAS30902.1 GEHC01000431 JAV47214.1 GGFK01012502 MBW45823.1 ATLV01027105 KE525423 KFB53967.1 KC740721 AGM32545.1 GFDL01001019 JAV34026.1 GFDL01001014 JAV34031.1 GFDL01001102 JAV33943.1 GECZ01024574 JAS45195.1 GGFJ01009527 MBW58668.1 ADMH02001585 ETN61969.1 AXCN02001078 KK852415 KDR24376.1 AXCM01002417 GGFM01008043 MBW28794.1 APCN01003273 AAAB01008986 EAA43293.4 GFDL01001263 JAV33782.1 GFDL01000988 JAV34057.1 UFQT01001137 SSX29101.1 GFDL01001274 JAV33771.1 GFDL01001297 JAV33748.1 KZ288229 PBC31663.1 GANO01003290 JAB56581.1 KQ414667 KOC64851.1 GDIQ01086670 JAN08067.1 GDIQ01132057 JAL19669.1 GDIP01081961 JAM21754.1 GDIP01079659 JAM24056.1 JH431251 GDIQ01155794 JAK95931.1 GDIP01156332 JAJ67070.1 GDIP01154809 JAJ68593.1 KQ759828 OAD62660.1 GL732537 EFX83319.1 AJVK01025330 GBBI01000679 JAC18033.1 GFTR01003005 JAW13421.1 GBGD01003146 JAC85743.1 GECL01000393 JAP05731.1 GDKW01000864 JAI55731.1 ACPB03024258 GAHY01001206 JAA76304.1 JH818919 EKC38196.1 GL443983 EFN61403.1 GL888463 EGI60601.1 NNAY01001930 OXU22525.1 KQ981727 KYN37190.1 KQ979120 KYN22571.1 KQ977565 KYN01947.1 ADTU01011664 KK107109 QOIP01000008 EZA59130.1 RLU19369.1 GBHO01045069 GDHC01018714 JAF98534.1 JAP99914.1 GDIQ01153045 JAK98680.1 KQ434809 KZC06584.1 GL764129 EFZ18477.1 KK116068 KFM66719.1 GDIQ01197175 JAK54550.1 GDIP01121554 JAL82160.1 GDIQ01193675 JAK58050.1 GL445814 EFN89118.1 GFDF01004545 JAV09539.1 GDIP01196199 JAJ27203.1 DS235006 EEB10380.1 AJWK01020518 NEDP02004821 OWF44352.1 GDIP01009434 JAM94281.1 GFJQ02002865 JAW04105.1 GGLE01002171 MBY06297.1 GFAA01000378 JAU03057.1 KQ422664 KOF74449.1 KQ435697 KOX80786.1 GBZX01002128 JAG90612.1 JH001110 EGW10943.1
PZC73292.1 GDQN01005433 JAT85621.1 KQ459465 KPJ00328.1 AGBW02014024 OWR42382.1 KQ460393 KPJ15357.1 PYGN01001693 PSN33336.1 NEVH01016292 PNF26151.1 GEDC01015014 GEDC01005778 JAS22284.1 JAS31520.1 CH477828 EAT35862.1 GEDC01006396 JAS30902.1 GEHC01000431 JAV47214.1 GGFK01012502 MBW45823.1 ATLV01027105 KE525423 KFB53967.1 KC740721 AGM32545.1 GFDL01001019 JAV34026.1 GFDL01001014 JAV34031.1 GFDL01001102 JAV33943.1 GECZ01024574 JAS45195.1 GGFJ01009527 MBW58668.1 ADMH02001585 ETN61969.1 AXCN02001078 KK852415 KDR24376.1 AXCM01002417 GGFM01008043 MBW28794.1 APCN01003273 AAAB01008986 EAA43293.4 GFDL01001263 JAV33782.1 GFDL01000988 JAV34057.1 UFQT01001137 SSX29101.1 GFDL01001274 JAV33771.1 GFDL01001297 JAV33748.1 KZ288229 PBC31663.1 GANO01003290 JAB56581.1 KQ414667 KOC64851.1 GDIQ01086670 JAN08067.1 GDIQ01132057 JAL19669.1 GDIP01081961 JAM21754.1 GDIP01079659 JAM24056.1 JH431251 GDIQ01155794 JAK95931.1 GDIP01156332 JAJ67070.1 GDIP01154809 JAJ68593.1 KQ759828 OAD62660.1 GL732537 EFX83319.1 AJVK01025330 GBBI01000679 JAC18033.1 GFTR01003005 JAW13421.1 GBGD01003146 JAC85743.1 GECL01000393 JAP05731.1 GDKW01000864 JAI55731.1 ACPB03024258 GAHY01001206 JAA76304.1 JH818919 EKC38196.1 GL443983 EFN61403.1 GL888463 EGI60601.1 NNAY01001930 OXU22525.1 KQ981727 KYN37190.1 KQ979120 KYN22571.1 KQ977565 KYN01947.1 ADTU01011664 KK107109 QOIP01000008 EZA59130.1 RLU19369.1 GBHO01045069 GDHC01018714 JAF98534.1 JAP99914.1 GDIQ01153045 JAK98680.1 KQ434809 KZC06584.1 GL764129 EFZ18477.1 KK116068 KFM66719.1 GDIQ01197175 JAK54550.1 GDIP01121554 JAL82160.1 GDIQ01193675 JAK58050.1 GL445814 EFN89118.1 GFDF01004545 JAV09539.1 GDIP01196199 JAJ27203.1 DS235006 EEB10380.1 AJWK01020518 NEDP02004821 OWF44352.1 GDIP01009434 JAM94281.1 GFJQ02002865 JAW04105.1 GGLE01002171 MBY06297.1 GFAA01000378 JAU03057.1 KQ422664 KOF74449.1 KQ435697 KOX80786.1 GBZX01002128 JAG90612.1 JH001110 EGW10943.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000245037
+ More
UP000235965 UP000008820 UP000075881 UP000030765 UP000075884 UP000075880 UP000000673 UP000075886 UP000076408 UP000027135 UP000075900 UP000075883 UP000075920 UP000075885 UP000069272 UP000076407 UP000075840 UP000075903 UP000007062 UP000002358 UP000242457 UP000005203 UP000053825 UP000000305 UP000092462 UP000015103 UP000005408 UP000000311 UP000007755 UP000215335 UP000078541 UP000078492 UP000078542 UP000005205 UP000053097 UP000279307 UP000085678 UP000076502 UP000054359 UP000075902 UP000008237 UP000009046 UP000092461 UP000242188 UP000007646 UP000053454 UP000053105 UP000001075
UP000235965 UP000008820 UP000075881 UP000030765 UP000075884 UP000075880 UP000000673 UP000075886 UP000076408 UP000027135 UP000075900 UP000075883 UP000075920 UP000075885 UP000069272 UP000076407 UP000075840 UP000075903 UP000007062 UP000002358 UP000242457 UP000005203 UP000053825 UP000000305 UP000092462 UP000015103 UP000005408 UP000000311 UP000007755 UP000215335 UP000078541 UP000078492 UP000078542 UP000005205 UP000053097 UP000279307 UP000085678 UP000076502 UP000054359 UP000075902 UP000008237 UP000009046 UP000092461 UP000242188 UP000007646 UP000053454 UP000053105 UP000001075
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JVC3
A0A2H1VI57
A0A2A4JL87
A0A2W1BDL2
A0A1E1WFH9
A0A194Q460
+ More
A0A212ELL7 A0A0N0PCZ1 A0A2P8XMW8 A0A2J7QC45 A0A1B6D9C5 Q16ND7 A0A1B6DYY7 A0A1W7R7U1 A0A2M4AYF4 A0A182K9U4 A0A084WUS3 R4UN35 A0A1Q3G2P1 A0A1Q3G2M3 A0A182N2L3 A0A1Q3G2B6 A0A1B6F4U1 A0A2M4C0Z9 A0A182IKF2 W5JGP6 A0A182QWZ2 A0A182Y219 A0A067RKI0 A0A182RYR4 A0A182MS10 A0A182W9D9 A0A2M3ZJT1 A0A182P1S5 A0A182FKM7 A0A182WZY1 A0A182HPU7 A0A182VDZ4 A0T4G7 A0A1Q3G1V9 A0A1Q3G2L0 A0A336MI34 A0A1Q3G1W8 A0A1Q3G1P4 K7IR81 A0A2A3EIP1 A0A088A625 U5ESE0 A0A0L7R253 A0A0P6CYD8 A0A0P5QNP6 A0A0P5WTM9 A0A0P5WZI5 T1IP91 A0A0P5MJG0 A0A0P5DH13 A0A0N8AEP1 A0A310SSR5 E9GAU8 A0A1B0D5J1 A0A023FAT8 A0A224XZC7 A0A069DNV1 A0A0V0GBZ4 A0A0P4VYN8 R4G407 K1RU74 E2AYP0 F4WYU7 A0A232EVY7 A0A195F9B2 A0A151JBN7 A0A195CN95 A0A158NC59 A0A026WSW3 A0A0A9W0Y3 A0A1S3IHD5 A0A0P5N8D4 A0A154P5H3 E9IM20 A0A087TNN0 A0A0P5JK99 A0A0P5V2H9 A0A0N8BBM3 A0A182UCT1 E2B5C3 A0A1L8DSV9 A0A0P5ANV1 E0VAH4 A0A1B0CNM0 A0A210Q6I6 A0A0P6CF42 A0A1Z5L9W8 A0A2R5LA70 A0A1E1XVB2 G3TCR7 A0A0L8GC71 A0A0M9AC34 A0A0C9SB84 G3I238
A0A212ELL7 A0A0N0PCZ1 A0A2P8XMW8 A0A2J7QC45 A0A1B6D9C5 Q16ND7 A0A1B6DYY7 A0A1W7R7U1 A0A2M4AYF4 A0A182K9U4 A0A084WUS3 R4UN35 A0A1Q3G2P1 A0A1Q3G2M3 A0A182N2L3 A0A1Q3G2B6 A0A1B6F4U1 A0A2M4C0Z9 A0A182IKF2 W5JGP6 A0A182QWZ2 A0A182Y219 A0A067RKI0 A0A182RYR4 A0A182MS10 A0A182W9D9 A0A2M3ZJT1 A0A182P1S5 A0A182FKM7 A0A182WZY1 A0A182HPU7 A0A182VDZ4 A0T4G7 A0A1Q3G1V9 A0A1Q3G2L0 A0A336MI34 A0A1Q3G1W8 A0A1Q3G1P4 K7IR81 A0A2A3EIP1 A0A088A625 U5ESE0 A0A0L7R253 A0A0P6CYD8 A0A0P5QNP6 A0A0P5WTM9 A0A0P5WZI5 T1IP91 A0A0P5MJG0 A0A0P5DH13 A0A0N8AEP1 A0A310SSR5 E9GAU8 A0A1B0D5J1 A0A023FAT8 A0A224XZC7 A0A069DNV1 A0A0V0GBZ4 A0A0P4VYN8 R4G407 K1RU74 E2AYP0 F4WYU7 A0A232EVY7 A0A195F9B2 A0A151JBN7 A0A195CN95 A0A158NC59 A0A026WSW3 A0A0A9W0Y3 A0A1S3IHD5 A0A0P5N8D4 A0A154P5H3 E9IM20 A0A087TNN0 A0A0P5JK99 A0A0P5V2H9 A0A0N8BBM3 A0A182UCT1 E2B5C3 A0A1L8DSV9 A0A0P5ANV1 E0VAH4 A0A1B0CNM0 A0A210Q6I6 A0A0P6CF42 A0A1Z5L9W8 A0A2R5LA70 A0A1E1XVB2 G3TCR7 A0A0L8GC71 A0A0M9AC34 A0A0C9SB84 G3I238
PDB
3FN1
E-value=6.45312e-60,
Score=580
Ontologies
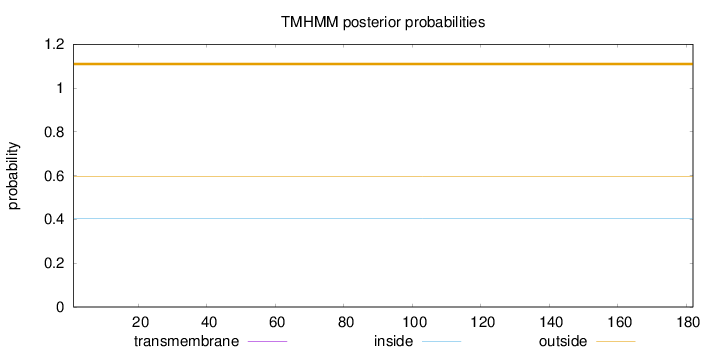
Topology
Length:
182
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00113
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.40472
outside
1 - 182
Population Genetic Test Statistics
Pi
253.49774
Theta
188.74862
Tajima's D
1.079535
CLR
0
CSRT
0.680265986700665
Interpretation
Uncertain
