Gene
KWMTBOMO02399
Pre Gene Modal
BGIBMGA013533
Annotation
PREDICTED:_dynein_assembly_factor_with_WDR_repeat_domains_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.592 Mitochondrial Reliability : 1.298
Sequence
CDS
ATGAAGTTATTGAAATTTCACCTTCGATATTACCCTCCGGGATTGATATTGGAATACTCGCAAAAAGGAACGATAAAGAATAAGGAGATTGATCTGTTGGATCTAAATACCAATTCAAACATAAAAGAAGTGGCACAGCATCTCTGCACCAAAGAGGCGTTAATCACGGACGAAGTGAGAGACCGATTGGAGCAATGTCTCGACACCTTAAAGAAGAGAATCGAACACGAAGGACCGGCTGGCAAAAGATTCTACATCTACAAGACATTACAGACACATTTATTGCCACTAACCAATGTGGCCTTTGACAAAACTGGGAAAAGATGTTTGACGGGCAGCTACGACAGAACTTGTAAGATATGGGACGTCGAAACCGGTAAGGAGCTAAAGAGCTTCTCCGGACATCAGAATGTCGTCTATTCAGTTGGCTTCAATTTTCCTGCCTGCGATCGCGTCCTAACGGGCTCCTTTGATAAAACTGCAAAAGTCTGGGATCCTTCTTCGGGTGAATGTCTGGCGACACTGTGGGGGCACCGAGCCGAAGTCGTGGCTGCTCAGTTCAGTTCCAAGGGAGACCTTGCTGCCACCGGCTCTATGGATCATACCGCTAAATTATTTGACTTAAAAACTGGTACCGAAATACACACATACAATGGTCACACAGCGGAGGTGGTCGCTCTGCAGTTTGATCCCAATGAAGGACGCCGTTTGATTACTGGATCCTTTGATGGTACCATCGCTCTGTGGGACGTTAGAGCTAAAGAGTTAGTATCACTTAATCTAAATAATATTACATCCACATAA
Protein
MKLLKFHLRYYPPGLILEYSQKGTIKNKEIDLLDLNTNSNIKEVAQHLCTKEALITDEVRDRLEQCLDTLKKRIEHEGPAGKRFYIYKTLQTHLLPLTNVAFDKTGKRCLTGSYDRTCKIWDVETGKELKSFSGHQNVVYSVGFNFPACDRVLTGSFDKTAKVWDPSSGECLATLWGHRAEVVAAQFSSKGDLAATGSMDHTAKLFDLKTGTEIHTYNGHTAEVVALQFDPNEGRRLITGSFDGTIALWDVRAKELVSLNLNNITST
Summary
Uniprot
H9JVH0
A0A2A4JTX5
A0A2W1BDJ1
A0A2H1VAA1
A0A212ELM1
A0A194QA91
+ More
A0A0N1PHM0 K7J563 A0A195F1A1 A0A0M8ZNI1 A0A195B2Q2 A0A0L7R3P4 A0A088AVV6 A0A195E6T5 A0A158NFY1 A0A182L5W6 A0A182WHL1 A0A182Y5N7 F4X6X2 E0VCG1 A0A182RRF7 A0A154PDZ8 A0A182PB24 A0A182NUZ6 A0A026VVY6 G4V7M5 A0A1S4FAH2 A0A067REJ0 A0A3M7QKS7 E2AMH6 A0A1B6KF52 A0A3Q0IS84 A0A151XC77 A0A1B6I6K7 A0A1B6FC70 A0A3Q0KI78 I7MMK9 C1BM69 A0A1U7RIR7 W5MNV1 A0A210PS13 R1BY77 A0A1S3SYB9 A0A3P9ACZ2 A0A3P9AE65 A0A336K8U9 A0A1R2CTK7 A0A1R2B4Z2 A0A0V0QQ61 W6L5J5 A0A182XCK5 A0A2C9JFA8 A0A1J1IZY9 A0A182IDY3 H2YPK6 A0A3L8D6Y1 A0A146XH47 A0A146XFZ2 A0A369S7K7 A0A1R2B2N8 F6UG25 A0A182M0T0 H3AA40 A0A2G8JYY1 K3X4I7 V3ZI80 A0A146XI61 G3W387 C5LLV7 A0A2D4BM24 A0A1W4YGW6 T1EF18 B3RLL7 A0A1R2BKP7 A0A2P4XD92 C3ZS55 G1KQC8 A0A0W8C2A9 A0A2G8K8K0 R7TY49 V9DZM1 W2VWN0 A0A3F2RPV6 W2PHU6 W2Y6Q2 A0A080Z309 W2HVY0 A0A329RY12 A0A2P4XFE0 A0A1W4Y770 A0A3B3T7G8 A0A225VM33 A0A1Q9C8A8 A0A1W4YGK9 H3GA86 A7SN68 G4ZNT8 F6TGM8 S9VIC3 K7FA48
A0A0N1PHM0 K7J563 A0A195F1A1 A0A0M8ZNI1 A0A195B2Q2 A0A0L7R3P4 A0A088AVV6 A0A195E6T5 A0A158NFY1 A0A182L5W6 A0A182WHL1 A0A182Y5N7 F4X6X2 E0VCG1 A0A182RRF7 A0A154PDZ8 A0A182PB24 A0A182NUZ6 A0A026VVY6 G4V7M5 A0A1S4FAH2 A0A067REJ0 A0A3M7QKS7 E2AMH6 A0A1B6KF52 A0A3Q0IS84 A0A151XC77 A0A1B6I6K7 A0A1B6FC70 A0A3Q0KI78 I7MMK9 C1BM69 A0A1U7RIR7 W5MNV1 A0A210PS13 R1BY77 A0A1S3SYB9 A0A3P9ACZ2 A0A3P9AE65 A0A336K8U9 A0A1R2CTK7 A0A1R2B4Z2 A0A0V0QQ61 W6L5J5 A0A182XCK5 A0A2C9JFA8 A0A1J1IZY9 A0A182IDY3 H2YPK6 A0A3L8D6Y1 A0A146XH47 A0A146XFZ2 A0A369S7K7 A0A1R2B2N8 F6UG25 A0A182M0T0 H3AA40 A0A2G8JYY1 K3X4I7 V3ZI80 A0A146XI61 G3W387 C5LLV7 A0A2D4BM24 A0A1W4YGW6 T1EF18 B3RLL7 A0A1R2BKP7 A0A2P4XD92 C3ZS55 G1KQC8 A0A0W8C2A9 A0A2G8K8K0 R7TY49 V9DZM1 W2VWN0 A0A3F2RPV6 W2PHU6 W2Y6Q2 A0A080Z309 W2HVY0 A0A329RY12 A0A2P4XFE0 A0A1W4Y770 A0A3B3T7G8 A0A225VM33 A0A1Q9C8A8 A0A1W4YGK9 H3GA86 A7SN68 G4ZNT8 F6TGM8 S9VIC3 K7FA48
Pubmed
19121390
28756777
22118469
26354079
20075255
21347285
+ More
20966253 25244985 21719571 20566863 24508170 22253936 17510324 24845553 30375419 20798317 16933976 28812685 23760476 25069045 26486372 24516393 15562597 30249741 30042472 17495919 9215903 29023486 23254933 21709235 18719581 28186564 18563158 29240929 16946064 17615350 19892987 23560078 17381049
20966253 25244985 21719571 20566863 24508170 22253936 17510324 24845553 30375419 20798317 16933976 28812685 23760476 25069045 26486372 24516393 15562597 30249741 30042472 17495919 9215903 29023486 23254933 21709235 18719581 28186564 18563158 29240929 16946064 17615350 19892987 23560078 17381049
EMBL
BABH01036149
BABH01036150
NWSH01000594
PCG75461.1
KZ150114
PZC73289.1
+ More
ODYU01001302 SOQ37322.1 AGBW02014024 OWR42385.1 KQ459465 KPJ00331.1 KQ460731 KPJ12642.1 AAZX01005033 KQ981864 KYN34243.1 KQ435969 KOX67900.1 KQ976662 KYM78560.1 KQ414663 KOC65510.1 KQ979568 KYN20893.1 ADTU01014832 GL888818 EGI57904.1 DS235053 EEB11067.1 KQ434870 KZC09498.1 KK107770 EZA47830.1 HE601624 CCD74941.1 KK852554 KDR21463.1 REGN01005919 RNA11575.1 GL440813 EFN65400.1 GEBQ01029932 JAT10045.1 KQ982314 KYQ57898.1 GECU01025176 JAS82530.1 GECZ01021963 JAS47806.1 GG662407 EAS05106.2 BT075698 ACO10122.1 AHAT01034251 NEDP02005537 OWF39244.1 KB867099 EOD15148.1 UFQS01000123 UFQT01000123 SSX00068.1 SSX20448.1 MPUH01000064 OMJ92328.1 MPUH01000950 OMJ71853.1 LDAU01000119 KRX04321.1 HF955091 CCW64776.1 CVRI01000065 CRL05773.1 APCN01004540 QOIP01000012 RLU16215.1 GCES01045394 JAR40929.1 GCES01045393 JAR40930.1 NOWV01000063 RDD42013.1 MPUH01001030 OMJ71019.1 AXCM01012117 AFYH01044252 AFYH01044253 AFYH01044254 AFYH01044255 MRZV01001074 PIK40905.1 GL376601 KB203567 ESO83902.1 GCES01045391 JAR40932.1 AEFK01135342 AEFK01135343 AEFK01135344 AEFK01135345 GG683299 EER02329.1 BBXB01000033 GAX95391.1 AMQM01003418 KB096134 ESO08141.1 DS985241 EDV28804.1 MPUH01000588 OMJ77195.1 NCKW01011429 POM63522.1 GG666670 EEN44634.1 LNFO01005408 KUF78164.1 MRZV01000785 PIK44295.1 AMQN01011376 KB308855 ELT96346.1 ANIZ01003664 ETI32289.1 ANIX01004139 ETP02168.1 MAYM02000837 MBDO02000138 MBAD02000682 RLN32379.1 RLN61942.1 RLN64084.1 KI669640 ETN00226.1 ANIY01004294 ETP30313.1 ANJA01003844 ETO61020.1 KI689532 KI676330 KI683061 KI696291 ETK72661.1 ETL26114.1 ETL79323.1 ETM32583.1 MJFZ01001227 MJFZ01000512 MJFZ01000195 RAW22727.1 RAW28108.1 RAW34617.1 NCKW01011146 POM64259.1 NBNE01003983 OWZ06392.1 LSRX01001525 OLP79107.1 DS566073 DS469717 EDO34865.1 JH159155 EGZ15406.1 ATMH01007999 EPY22960.1 AGCU01063966 AGCU01063967 AGCU01063968 AGCU01063969 AGCU01063970 AGCU01063971 AGCU01063972 AGCU01063973 AGCU01063974
ODYU01001302 SOQ37322.1 AGBW02014024 OWR42385.1 KQ459465 KPJ00331.1 KQ460731 KPJ12642.1 AAZX01005033 KQ981864 KYN34243.1 KQ435969 KOX67900.1 KQ976662 KYM78560.1 KQ414663 KOC65510.1 KQ979568 KYN20893.1 ADTU01014832 GL888818 EGI57904.1 DS235053 EEB11067.1 KQ434870 KZC09498.1 KK107770 EZA47830.1 HE601624 CCD74941.1 KK852554 KDR21463.1 REGN01005919 RNA11575.1 GL440813 EFN65400.1 GEBQ01029932 JAT10045.1 KQ982314 KYQ57898.1 GECU01025176 JAS82530.1 GECZ01021963 JAS47806.1 GG662407 EAS05106.2 BT075698 ACO10122.1 AHAT01034251 NEDP02005537 OWF39244.1 KB867099 EOD15148.1 UFQS01000123 UFQT01000123 SSX00068.1 SSX20448.1 MPUH01000064 OMJ92328.1 MPUH01000950 OMJ71853.1 LDAU01000119 KRX04321.1 HF955091 CCW64776.1 CVRI01000065 CRL05773.1 APCN01004540 QOIP01000012 RLU16215.1 GCES01045394 JAR40929.1 GCES01045393 JAR40930.1 NOWV01000063 RDD42013.1 MPUH01001030 OMJ71019.1 AXCM01012117 AFYH01044252 AFYH01044253 AFYH01044254 AFYH01044255 MRZV01001074 PIK40905.1 GL376601 KB203567 ESO83902.1 GCES01045391 JAR40932.1 AEFK01135342 AEFK01135343 AEFK01135344 AEFK01135345 GG683299 EER02329.1 BBXB01000033 GAX95391.1 AMQM01003418 KB096134 ESO08141.1 DS985241 EDV28804.1 MPUH01000588 OMJ77195.1 NCKW01011429 POM63522.1 GG666670 EEN44634.1 LNFO01005408 KUF78164.1 MRZV01000785 PIK44295.1 AMQN01011376 KB308855 ELT96346.1 ANIZ01003664 ETI32289.1 ANIX01004139 ETP02168.1 MAYM02000837 MBDO02000138 MBAD02000682 RLN32379.1 RLN61942.1 RLN64084.1 KI669640 ETN00226.1 ANIY01004294 ETP30313.1 ANJA01003844 ETO61020.1 KI689532 KI676330 KI683061 KI696291 ETK72661.1 ETL26114.1 ETL79323.1 ETM32583.1 MJFZ01001227 MJFZ01000512 MJFZ01000195 RAW22727.1 RAW28108.1 RAW34617.1 NCKW01011146 POM64259.1 NBNE01003983 OWZ06392.1 LSRX01001525 OLP79107.1 DS566073 DS469717 EDO34865.1 JH159155 EGZ15406.1 ATMH01007999 EPY22960.1 AGCU01063966 AGCU01063967 AGCU01063968 AGCU01063969 AGCU01063970 AGCU01063971 AGCU01063972 AGCU01063973 AGCU01063974
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000002358
+ More
UP000078541 UP000053105 UP000078540 UP000053825 UP000005203 UP000078492 UP000005205 UP000075882 UP000075920 UP000076408 UP000007755 UP000009046 UP000075900 UP000076502 UP000075885 UP000075884 UP000053097 UP000008854 UP000027135 UP000276133 UP000000311 UP000079169 UP000075809 UP000009168 UP000189705 UP000018468 UP000242188 UP000013827 UP000087266 UP000265140 UP000076407 UP000076420 UP000183832 UP000075840 UP000007875 UP000279307 UP000253843 UP000002280 UP000075883 UP000008672 UP000230750 UP000030746 UP000007648 UP000054177 UP000192224 UP000015101 UP000009022 UP000001554 UP000001646 UP000052943 UP000014760 UP000018721 UP000018958 UP000277300 UP000284657 UP000285883 UP000018817 UP000018948 UP000028582 UP000053236 UP000053864 UP000054423 UP000054532 UP000251314 UP000261540 UP000198211 UP000005238 UP000001593 UP000002640 UP000002281 UP000007267
UP000078541 UP000053105 UP000078540 UP000053825 UP000005203 UP000078492 UP000005205 UP000075882 UP000075920 UP000076408 UP000007755 UP000009046 UP000075900 UP000076502 UP000075885 UP000075884 UP000053097 UP000008854 UP000027135 UP000276133 UP000000311 UP000079169 UP000075809 UP000009168 UP000189705 UP000018468 UP000242188 UP000013827 UP000087266 UP000265140 UP000076407 UP000076420 UP000183832 UP000075840 UP000007875 UP000279307 UP000253843 UP000002280 UP000075883 UP000008672 UP000230750 UP000030746 UP000007648 UP000054177 UP000192224 UP000015101 UP000009022 UP000001554 UP000001646 UP000052943 UP000014760 UP000018721 UP000018958 UP000277300 UP000284657 UP000285883 UP000018817 UP000018948 UP000028582 UP000053236 UP000053864 UP000054423 UP000054532 UP000251314 UP000261540 UP000198211 UP000005238 UP000001593 UP000002640 UP000002281 UP000007267
PRIDE
Interpro
IPR001680
WD40_repeat
+ More
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR011047 Quinoprotein_ADH-like_supfam
IPR011044 Quino_amine_DH_bsu
IPR011043 Gal_Oxase/kelch_b-propeller
IPR029412 CEP19
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR002350 Kazal_dom
IPR004156 OATP
IPR036058 Kazal_dom_sf
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR011047 Quinoprotein_ADH-like_supfam
IPR011044 Quino_amine_DH_bsu
IPR011043 Gal_Oxase/kelch_b-propeller
IPR029412 CEP19
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR002350 Kazal_dom
IPR004156 OATP
IPR036058 Kazal_dom_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JVH0
A0A2A4JTX5
A0A2W1BDJ1
A0A2H1VAA1
A0A212ELM1
A0A194QA91
+ More
A0A0N1PHM0 K7J563 A0A195F1A1 A0A0M8ZNI1 A0A195B2Q2 A0A0L7R3P4 A0A088AVV6 A0A195E6T5 A0A158NFY1 A0A182L5W6 A0A182WHL1 A0A182Y5N7 F4X6X2 E0VCG1 A0A182RRF7 A0A154PDZ8 A0A182PB24 A0A182NUZ6 A0A026VVY6 G4V7M5 A0A1S4FAH2 A0A067REJ0 A0A3M7QKS7 E2AMH6 A0A1B6KF52 A0A3Q0IS84 A0A151XC77 A0A1B6I6K7 A0A1B6FC70 A0A3Q0KI78 I7MMK9 C1BM69 A0A1U7RIR7 W5MNV1 A0A210PS13 R1BY77 A0A1S3SYB9 A0A3P9ACZ2 A0A3P9AE65 A0A336K8U9 A0A1R2CTK7 A0A1R2B4Z2 A0A0V0QQ61 W6L5J5 A0A182XCK5 A0A2C9JFA8 A0A1J1IZY9 A0A182IDY3 H2YPK6 A0A3L8D6Y1 A0A146XH47 A0A146XFZ2 A0A369S7K7 A0A1R2B2N8 F6UG25 A0A182M0T0 H3AA40 A0A2G8JYY1 K3X4I7 V3ZI80 A0A146XI61 G3W387 C5LLV7 A0A2D4BM24 A0A1W4YGW6 T1EF18 B3RLL7 A0A1R2BKP7 A0A2P4XD92 C3ZS55 G1KQC8 A0A0W8C2A9 A0A2G8K8K0 R7TY49 V9DZM1 W2VWN0 A0A3F2RPV6 W2PHU6 W2Y6Q2 A0A080Z309 W2HVY0 A0A329RY12 A0A2P4XFE0 A0A1W4Y770 A0A3B3T7G8 A0A225VM33 A0A1Q9C8A8 A0A1W4YGK9 H3GA86 A7SN68 G4ZNT8 F6TGM8 S9VIC3 K7FA48
A0A0N1PHM0 K7J563 A0A195F1A1 A0A0M8ZNI1 A0A195B2Q2 A0A0L7R3P4 A0A088AVV6 A0A195E6T5 A0A158NFY1 A0A182L5W6 A0A182WHL1 A0A182Y5N7 F4X6X2 E0VCG1 A0A182RRF7 A0A154PDZ8 A0A182PB24 A0A182NUZ6 A0A026VVY6 G4V7M5 A0A1S4FAH2 A0A067REJ0 A0A3M7QKS7 E2AMH6 A0A1B6KF52 A0A3Q0IS84 A0A151XC77 A0A1B6I6K7 A0A1B6FC70 A0A3Q0KI78 I7MMK9 C1BM69 A0A1U7RIR7 W5MNV1 A0A210PS13 R1BY77 A0A1S3SYB9 A0A3P9ACZ2 A0A3P9AE65 A0A336K8U9 A0A1R2CTK7 A0A1R2B4Z2 A0A0V0QQ61 W6L5J5 A0A182XCK5 A0A2C9JFA8 A0A1J1IZY9 A0A182IDY3 H2YPK6 A0A3L8D6Y1 A0A146XH47 A0A146XFZ2 A0A369S7K7 A0A1R2B2N8 F6UG25 A0A182M0T0 H3AA40 A0A2G8JYY1 K3X4I7 V3ZI80 A0A146XI61 G3W387 C5LLV7 A0A2D4BM24 A0A1W4YGW6 T1EF18 B3RLL7 A0A1R2BKP7 A0A2P4XD92 C3ZS55 G1KQC8 A0A0W8C2A9 A0A2G8K8K0 R7TY49 V9DZM1 W2VWN0 A0A3F2RPV6 W2PHU6 W2Y6Q2 A0A080Z309 W2HVY0 A0A329RY12 A0A2P4XFE0 A0A1W4Y770 A0A3B3T7G8 A0A225VM33 A0A1Q9C8A8 A0A1W4YGK9 H3GA86 A7SN68 G4ZNT8 F6TGM8 S9VIC3 K7FA48
Ontologies
GO
PANTHER
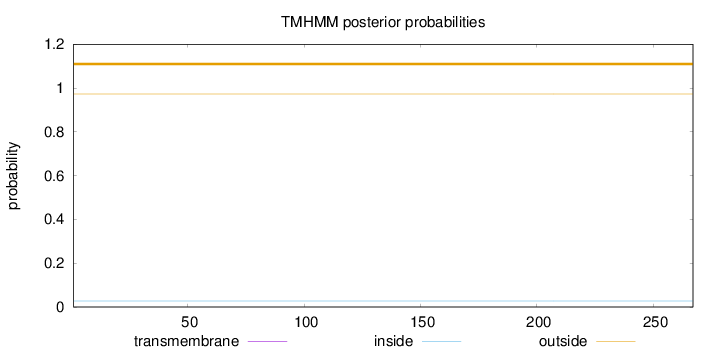
Topology
Length:
267
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00101
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02767
outside
1 - 267
Population Genetic Test Statistics
Pi
164.766962
Theta
155.945938
Tajima's D
0.120817
CLR
0.318796
CSRT
0.395930203489825
Interpretation
Possibly Positive selection
