Gene
KWMTBOMO02391
Annotation
PREDICTED:_mutS_protein_homolog_5-like?_partial_[Plutella_xylostella]
Full name
DNA mismatch repair protein
Location in the cell
PlasmaMembrane Reliability : 4.009
Sequence
CDS
ATGTCGCTTACATTCCACCACACGGGCAGTTCACAGTATCAAAAGTTCGGTAGGGCAATCAGCCAATTGACCAGGAGCATAGTTGTTAAAACATTATTAATATCTCCAGGTCTGATAGTGTATTTAGCTCACATCGGTAGCTTCGTGCCGGCTGACTCGGCTGTCATAGGCATCGTGAGTCACATATTCACAAGGATGCAGTCAACTGAATGCATCGCAGCGCACATGTCAGCTTTCCTAATAGATTTGAGACAGATGGCTCTGGTTCTACAAGAGTCGACGAGTAGCAGCCTGGTGCTGGTGGACGAGTTCGGCAAAGGCACAGCGCGGCGCGACGGGGCGGCCCTGCTGGCGGCCGCGCTGGGCGCCGTGCTGGCTCGCGCGCGCCGCCCGCACGTGCTGCTCGCCACGCACTTCCTGCAGCTGCACCACTACATCGCTCCCTCGCCACTACTCACGTTCTTGGTAATATTGACAACGCCATTCTTATAG
Protein
MSLTFHHTGSSQYQKFGRAISQLTRSIVVKTLLISPGLIVYLAHIGSFVPADSAVIGIVSHIFTRMQSTECIAAHMSAFLIDLRQMALVLQESTSSSLVLVDEFGKGTARRDGAALLAAALGAVLARARRPHVLLATHFLQLHHYIAPSPLLTFLVILTTPFL
Summary
Description
Component of the post-replicative DNA mismatch repair system (MMR).
Similarity
Belongs to the DNA mismatch repair MutS family.
Uniprot
A0A194Q4R3
A0A0N1IDJ9
A0A2H1WF92
A0A2A4JH95
A0A1B6ELS4
A0A146S8D9
+ More
A0A3B3SSG7 A0A146TVZ6 T1KLA5 A0A146RXM7 A0A2S2NX47 A0A044U217 A0A0V1IFF7 A0A0V1IFG5 A0A0V1FV26 A0A0V0XWS9 A0A0V0XWM4 A0A182EPI0 A0A0V0XWQ3 A0A0V0XXA2 A0A0V0XWC5 A0A0R3S1D7 A0A0R3QYZ0 A0A1A8C3W8 A0A0V1JCF7 A0A0V1JCH7 A0A0V1JDR0 A0A0V1JCH6 A0A0V1JCP0 A0A0V1JCF3 A0A0V1JCL3 A0A0V1JCH1 A0A3P6U166 A0A158PYW5 A0A1I9G7E3 A0A0N4TRY3 A0A3B3XQX1 A0A183H916 A0A2R6WVC8 A0A2R6WVB2 V4AGJ6 A0A210PVY1 A0A2R6WVC2 A0A2R6WVC4 A0A3B5N002 A0A1S0UGB4 T1ETZ2 A0A315WBZ4 A0A3P9PXH0 A0A1I7VHQ1 A0A1A8QLR5 A0A2S2QKF9 J9LB75 A0A2R6WVC0 A0A238C477 A0A2R6WVC5 A0A0V1N1Y6 A0A0V1HY88 A0A1A8AJJ1 A0A1A7ZE46 A0A158R5J7 A0A1B6KRA6 A0A3B4BAL3 A0A3P8RTP1 A0A0K0MXG8 A0A0V0RZG1 A0A0V0RZK2 A0A0V0RZU3 A0A2J6SCK3 A0A3M6UXS0 J9EWB9 A0A3P7DED9 A0A1B6CPF5 A0A3P8RUS4 A0A0G2USR8 A7EN13 A0A1D9QID6 F6YKR1 A0A2B4SNF7
A0A3B3SSG7 A0A146TVZ6 T1KLA5 A0A146RXM7 A0A2S2NX47 A0A044U217 A0A0V1IFF7 A0A0V1IFG5 A0A0V1FV26 A0A0V0XWS9 A0A0V0XWM4 A0A182EPI0 A0A0V0XWQ3 A0A0V0XXA2 A0A0V0XWC5 A0A0R3S1D7 A0A0R3QYZ0 A0A1A8C3W8 A0A0V1JCF7 A0A0V1JCH7 A0A0V1JDR0 A0A0V1JCH6 A0A0V1JCP0 A0A0V1JCF3 A0A0V1JCL3 A0A0V1JCH1 A0A3P6U166 A0A158PYW5 A0A1I9G7E3 A0A0N4TRY3 A0A3B3XQX1 A0A183H916 A0A2R6WVC8 A0A2R6WVB2 V4AGJ6 A0A210PVY1 A0A2R6WVC2 A0A2R6WVC4 A0A3B5N002 A0A1S0UGB4 T1ETZ2 A0A315WBZ4 A0A3P9PXH0 A0A1I7VHQ1 A0A1A8QLR5 A0A2S2QKF9 J9LB75 A0A2R6WVC0 A0A238C477 A0A2R6WVC5 A0A0V1N1Y6 A0A0V1HY88 A0A1A8AJJ1 A0A1A7ZE46 A0A158R5J7 A0A1B6KRA6 A0A3B4BAL3 A0A3P8RTP1 A0A0K0MXG8 A0A0V0RZG1 A0A0V0RZK2 A0A0V0RZU3 A0A2J6SCK3 A0A3M6UXS0 J9EWB9 A0A3P7DED9 A0A1B6CPF5 A0A3P8RUS4 A0A0G2USR8 A7EN13 A0A1D9QID6 F6YKR1 A0A2B4SNF7
Pubmed
EMBL
KQ459465
KPJ00339.1
KQ460731
KPJ12649.1
ODYU01008277
SOQ51721.1
+ More
NWSH01001581 PCG70800.1 GECZ01030946 JAS38823.1 GCES01109896 JAQ76426.1 GCES01089091 JAQ97231.1 CAEY01000211 GCES01113233 JAQ73089.1 GGMR01009174 MBY21793.1 CMVM020000166 JYDS01000202 KRZ21514.1 KRZ21515.1 JYDT01000028 KRY89713.1 JYDU01000114 KRX92357.1 KRX92359.1 UYRW01005370 VDM93755.1 KRX92362.1 KRX92361.1 KRX92360.1 UZAG01017954 VDO37445.1 HADZ01010328 SBP74269.1 JYDV01000109 KRZ32674.1 KRZ32672.1 KRZ32677.1 KRZ32673.1 KRZ32676.1 KRZ32678.1 KRZ32671.1 KRZ32675.1 UYRX01001012 VDK87685.1 LN856635 CDQ04605.1 UZAD01013228 VDN92582.1 UZAJ01002852 VDO38463.1 KZ772727 PTQ37796.1 PTQ37797.1 KB202050 ESO92536.1 NEDP02005458 OWF40626.1 PTQ37798.1 PTQ37799.1 JH712232 EJD74740.1 AMQM01001381 KB097495 ESN96325.1 NHOQ01000034 PWA33565.1 HAEG01013243 SBR94422.1 GGMS01009046 MBY78249.1 ABLF02037161 PTQ37801.1 KZ269978 OZC12272.1 PTQ37803.1 JYDO01000015 KRZ78056.1 JYDP01000017 KRZ15548.1 HADY01016368 SBP54853.1 HADY01002273 SBP40758.1 GEBQ01026010 JAT13967.1 KM283882 AKI32379.1 JYDL01000054 KRX19902.1 KRX19904.1 KRX19903.1 KZ613937 PMD48501.1 RCHS01000531 RMX58389.1 ADBV01000563 EJW86906.1 UYWW01000332 VDM08160.1 GEDC01021936 JAS15362.1 KP259582 AKI32545.1 CH476628 EDO04229.1 CP017826 APA14707.1 LSMT01000056 PFX30047.1
NWSH01001581 PCG70800.1 GECZ01030946 JAS38823.1 GCES01109896 JAQ76426.1 GCES01089091 JAQ97231.1 CAEY01000211 GCES01113233 JAQ73089.1 GGMR01009174 MBY21793.1 CMVM020000166 JYDS01000202 KRZ21514.1 KRZ21515.1 JYDT01000028 KRY89713.1 JYDU01000114 KRX92357.1 KRX92359.1 UYRW01005370 VDM93755.1 KRX92362.1 KRX92361.1 KRX92360.1 UZAG01017954 VDO37445.1 HADZ01010328 SBP74269.1 JYDV01000109 KRZ32674.1 KRZ32672.1 KRZ32677.1 KRZ32673.1 KRZ32676.1 KRZ32678.1 KRZ32671.1 KRZ32675.1 UYRX01001012 VDK87685.1 LN856635 CDQ04605.1 UZAD01013228 VDN92582.1 UZAJ01002852 VDO38463.1 KZ772727 PTQ37796.1 PTQ37797.1 KB202050 ESO92536.1 NEDP02005458 OWF40626.1 PTQ37798.1 PTQ37799.1 JH712232 EJD74740.1 AMQM01001381 KB097495 ESN96325.1 NHOQ01000034 PWA33565.1 HAEG01013243 SBR94422.1 GGMS01009046 MBY78249.1 ABLF02037161 PTQ37801.1 KZ269978 OZC12272.1 PTQ37803.1 JYDO01000015 KRZ78056.1 JYDP01000017 KRZ15548.1 HADY01016368 SBP54853.1 HADY01002273 SBP40758.1 GEBQ01026010 JAT13967.1 KM283882 AKI32379.1 JYDL01000054 KRX19902.1 KRX19904.1 KRX19903.1 KZ613937 PMD48501.1 RCHS01000531 RMX58389.1 ADBV01000563 EJW86906.1 UYWW01000332 VDM08160.1 GEDC01021936 JAS15362.1 KP259582 AKI32545.1 CH476628 EDO04229.1 CP017826 APA14707.1 LSMT01000056 PFX30047.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000261540
UP000015104
UP000024404
+ More
UP000054805 UP000054995 UP000054815 UP000077448 UP000271087 UP000050640 UP000050602 UP000054826 UP000277928 UP000006672 UP000038020 UP000278627 UP000261480 UP000050787 UP000030746 UP000242188 UP000261380 UP000015101 UP000242638 UP000095285 UP000007819 UP000054843 UP000055024 UP000046393 UP000261520 UP000265080 UP000054630 UP000235786 UP000275408 UP000004810 UP000270924 UP000001312 UP000177798 UP000008144 UP000225706
UP000054805 UP000054995 UP000054815 UP000077448 UP000271087 UP000050640 UP000050602 UP000054826 UP000277928 UP000006672 UP000038020 UP000278627 UP000261480 UP000050787 UP000030746 UP000242188 UP000261380 UP000015101 UP000242638 UP000095285 UP000007819 UP000054843 UP000055024 UP000046393 UP000261520 UP000265080 UP000054630 UP000235786 UP000275408 UP000004810 UP000270924 UP000001312 UP000177798 UP000008144 UP000225706
Pfam
Interpro
IPR000432
DNA_mismatch_repair_MutS_C
+ More
IPR027417 P-loop_NTPase
IPR036187 DNA_mismatch_repair_MutS_sf
IPR001612 Caveolin
IPR007696 DNA_mismatch_repair_MutS_core
IPR017261 DNA_mismatch_repair_MutS/MSH
IPR007861 DNA_mismatch_repair_MutS_clamp
IPR011184 DNA_mismatch_repair_Msh2
IPR030387 G_Bms1/Tsr1_dom
IPR036678 MutS_con_dom_sf
IPR013078 His_Pase_superF_clade-1
IPR029033 His_PPase_superfam
IPR000306 Znf_FYVE
IPR022557 DUF3480
IPR011011 Znf_FYVE_PHD
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR007860 DNA_mmatch_repair_MutS_con_dom
IPR027417 P-loop_NTPase
IPR036187 DNA_mismatch_repair_MutS_sf
IPR001612 Caveolin
IPR007696 DNA_mismatch_repair_MutS_core
IPR017261 DNA_mismatch_repair_MutS/MSH
IPR007861 DNA_mismatch_repair_MutS_clamp
IPR011184 DNA_mismatch_repair_Msh2
IPR030387 G_Bms1/Tsr1_dom
IPR036678 MutS_con_dom_sf
IPR013078 His_Pase_superF_clade-1
IPR029033 His_PPase_superfam
IPR000306 Znf_FYVE
IPR022557 DUF3480
IPR011011 Znf_FYVE_PHD
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR007860 DNA_mmatch_repair_MutS_con_dom
SUPFAM
Gene 3D
ProteinModelPortal
A0A194Q4R3
A0A0N1IDJ9
A0A2H1WF92
A0A2A4JH95
A0A1B6ELS4
A0A146S8D9
+ More
A0A3B3SSG7 A0A146TVZ6 T1KLA5 A0A146RXM7 A0A2S2NX47 A0A044U217 A0A0V1IFF7 A0A0V1IFG5 A0A0V1FV26 A0A0V0XWS9 A0A0V0XWM4 A0A182EPI0 A0A0V0XWQ3 A0A0V0XXA2 A0A0V0XWC5 A0A0R3S1D7 A0A0R3QYZ0 A0A1A8C3W8 A0A0V1JCF7 A0A0V1JCH7 A0A0V1JDR0 A0A0V1JCH6 A0A0V1JCP0 A0A0V1JCF3 A0A0V1JCL3 A0A0V1JCH1 A0A3P6U166 A0A158PYW5 A0A1I9G7E3 A0A0N4TRY3 A0A3B3XQX1 A0A183H916 A0A2R6WVC8 A0A2R6WVB2 V4AGJ6 A0A210PVY1 A0A2R6WVC2 A0A2R6WVC4 A0A3B5N002 A0A1S0UGB4 T1ETZ2 A0A315WBZ4 A0A3P9PXH0 A0A1I7VHQ1 A0A1A8QLR5 A0A2S2QKF9 J9LB75 A0A2R6WVC0 A0A238C477 A0A2R6WVC5 A0A0V1N1Y6 A0A0V1HY88 A0A1A8AJJ1 A0A1A7ZE46 A0A158R5J7 A0A1B6KRA6 A0A3B4BAL3 A0A3P8RTP1 A0A0K0MXG8 A0A0V0RZG1 A0A0V0RZK2 A0A0V0RZU3 A0A2J6SCK3 A0A3M6UXS0 J9EWB9 A0A3P7DED9 A0A1B6CPF5 A0A3P8RUS4 A0A0G2USR8 A7EN13 A0A1D9QID6 F6YKR1 A0A2B4SNF7
A0A3B3SSG7 A0A146TVZ6 T1KLA5 A0A146RXM7 A0A2S2NX47 A0A044U217 A0A0V1IFF7 A0A0V1IFG5 A0A0V1FV26 A0A0V0XWS9 A0A0V0XWM4 A0A182EPI0 A0A0V0XWQ3 A0A0V0XXA2 A0A0V0XWC5 A0A0R3S1D7 A0A0R3QYZ0 A0A1A8C3W8 A0A0V1JCF7 A0A0V1JCH7 A0A0V1JDR0 A0A0V1JCH6 A0A0V1JCP0 A0A0V1JCF3 A0A0V1JCL3 A0A0V1JCH1 A0A3P6U166 A0A158PYW5 A0A1I9G7E3 A0A0N4TRY3 A0A3B3XQX1 A0A183H916 A0A2R6WVC8 A0A2R6WVB2 V4AGJ6 A0A210PVY1 A0A2R6WVC2 A0A2R6WVC4 A0A3B5N002 A0A1S0UGB4 T1ETZ2 A0A315WBZ4 A0A3P9PXH0 A0A1I7VHQ1 A0A1A8QLR5 A0A2S2QKF9 J9LB75 A0A2R6WVC0 A0A238C477 A0A2R6WVC5 A0A0V1N1Y6 A0A0V1HY88 A0A1A8AJJ1 A0A1A7ZE46 A0A158R5J7 A0A1B6KRA6 A0A3B4BAL3 A0A3P8RTP1 A0A0K0MXG8 A0A0V0RZG1 A0A0V0RZK2 A0A0V0RZU3 A0A2J6SCK3 A0A3M6UXS0 J9EWB9 A0A3P7DED9 A0A1B6CPF5 A0A3P8RUS4 A0A0G2USR8 A7EN13 A0A1D9QID6 F6YKR1 A0A2B4SNF7
PDB
5YK4
E-value=1.03451e-13,
Score=181
Ontologies
GO
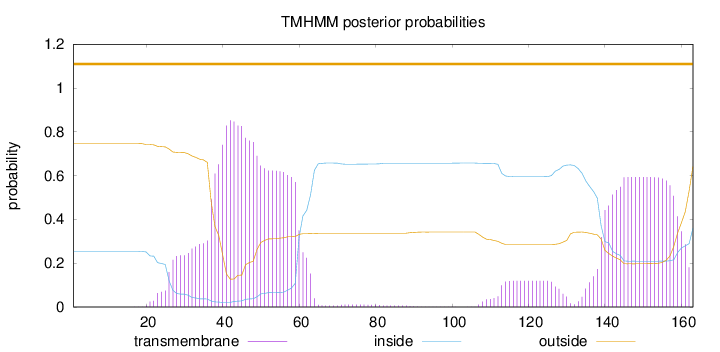
Topology
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
34.98273
Exp number, first 60 AAs:
19.16489
Total prob of N-in:
0.25401
POSSIBLE N-term signal
sequence
outside
1 - 163
Population Genetic Test Statistics
Pi
294.330841
Theta
173.934276
Tajima's D
1.654043
CLR
0.510377
CSRT
0.823408829558522
Interpretation
Uncertain
