Gene
KWMTBOMO02386
Pre Gene Modal
BGIBMGA013538
Annotation
PREDICTED:_bromodomain-containing_protein_7_[Bombyx_mori]
Full name
Serine hydroxymethyltransferase
Location in the cell
Nuclear Reliability : 4.288
Sequence
CDS
ATGGATGACATAGAAAAGCTGAGTTGCAGAGAAGAGATTTTAGAAATGAGTGATCTGGAGCACTCTGCAACCGACGATTTTAGAGAAACGGAAGGAAAATCAAAAAAGAAGAAAAGAAGACGCCGCGAAATAGATATGAACTCAGGGAAGCATATTTTTAAAAAGATAAAGCTGTCAGATGACGATGAGACAGATGATGAGATTATGATGAAAAAAATGATGCGGAACTTCAGCCCCGGACCTAACTCACCAGCTTCTCACAGAGAACCACGGACTTGTGTCCTGAAGGCTAGTCAAAAACGTCGTCCACTCTCCCGTTTGCTTGAACAGCTACTACGGAACTTAGAGAAAAGAGACCCACACCAGTTCTTTGCTTGGCCCGTCAATGATAACTTTGCTCCAGGCTACTCTACCATCATCAAGAAACCCATGGATTTCTCTACTATGAAGCAGAAGATTGATGATAATGAATACAAATCCCTTAATTGTTTTATTAGCGACTTCAAACTGATGTGCAACAATGCCATGAAGTACAACAAACCAGGCACCGTGTACCACAAGGCAGCCCGGAGACTCTTGCACGCCGGTCTCAAGCAGCTCACTCCGCAGAAACTGAGGCCGCTGGGAGACATCCTCACCTATATGTACGAGATACCCATTAGAGAGCTAGGATTCGACATGGGGAAAATGGATGTGCAGAGGGTGCTCAAACGGAACAGCCCCGTGAAGAGCGGGGGCTCCGAGCCCGAGGGCCCCGGGGACCCGGACTCCGTCAAGATACACATGGAAGCCGCGCGCGAGCAGCACCGCCGGAGACTCGCCAAGAAAGCCTTTCCGAGAATGGACGCGGAAGGGAAGACAACATTGTGCCTGGTCACCAACACGGTTGAGGGGAACGGTGAAGACAAACCCATGACCCTCGGAAAACTCATCGGGAAACTGAATCAGGGGACAGGAACTTTACACACTCCACGCGAAGACCGTCGCAACGTGGCGAAGGGTGTGAAGCCCTTGAACTACGGCCCCTTCAGCTCGTACGCGCCCTCCTACGACGGGACCTTCGCCACCCTCACCAAGGAGGAGTCGCACCTCATCTATCACACGCTACCATCAGAAACGGGACTAGGAAACGAGGAGATGCTGCGGTTCGCGCCGGACTCGCCCTGGGCCCTGCTCTACGACATGGAGGCGCTGTTCCCGGACAAGAAGCACGCGGCTGAGACCGCCCCCCCGCCTCACGACAACGAGCTGTCTAAAGTGAAGATAGACCTGGAGGAGTTGCGCTCGCTGTCGGCGCTGGGCGTGGACGTGGAGTTCGTGGCGGAGGCGGAGCGGGCCGCGCGCGACCTGGGGCTCGGGCCCGCGCTCGCGCACACCGCGCAGCTCTTGCAGCGGTTGCAGCACGAGCAGCACCAGCGGTAA
Protein
MDDIEKLSCREEILEMSDLEHSATDDFRETEGKSKKKKRRRREIDMNSGKHIFKKIKLSDDDETDDEIMMKKMMRNFSPGPNSPASHREPRTCVLKASQKRRPLSRLLEQLLRNLEKRDPHQFFAWPVNDNFAPGYSTIIKKPMDFSTMKQKIDDNEYKSLNCFISDFKLMCNNAMKYNKPGTVYHKAARRLLHAGLKQLTPQKLRPLGDILTYMYEIPIRELGFDMGKMDVQRVLKRNSPVKSGGSEPEGPGDPDSVKIHMEAAREQHRRRLAKKAFPRMDAEGKTTLCLVTNTVEGNGEDKPMTLGKLIGKLNQGTGTLHTPREDRRNVAKGVKPLNYGPFSSYAPSYDGTFATLTKEESHLIYHTLPSETGLGNEEMLRFAPDSPWALLYDMEALFPDKKHAAETAPPPHDNELSKVKIDLEELRSLSALGVDVEFVAEAERAARDLGLGPALAHTAQLLQRLQHEQHQR
Summary
Description
Interconversion of serine and glycine.
Catalytic Activity
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + glycine + H2O = (6S)-5,6,7,8-tetrahydrofolate + L-serine
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the SHMT family.
Uniprot
H9JVH5
A0A2H1VPZ8
A0A2W1BF18
S4PDX7
A0A212FH88
A0A194Q5R4
+ More
A0A0N1IPM5 A0A0L7L4J8 A0A0C9RUM2 A0A0C9RDX9 E2BEL8 A0A151WUU2 F4WAC0 A0A195CSG6 A0A158NKY0 A0A195FSA4 A0A195E454 E0VMG6 A0A154P2U9 A0A088A0B3 A0A2A3ES74 E9IZ18 A0A0J7L877 D2A3W1 A0A2J7RIK3 E2AL99 A0A067RX80 A0A026WME5 U5EPR5 A0A182HCX5 A0A1B0DRE2 A0A1B0DFM0 A0A0L7R1P3 A0A1Q3F2B7 A0A0A9Z2Y2 A0A0M9A3N5 A0A0K8SQJ5 A0A0A9Z6V7 A0A1B6DLS8 A0A182RAW2 A0A1Q3F3E8 A0A182Q0T5 A0A182P9G8 T1HMJ5 A0A182MZ24 A0A084W1X9 B0W8Y1 Q17J04 A0A1S4F196 A0A023EZU8 A0A0V0G908 W5J5K1 A0A069DXD8 A0A224XJV3 A0A182F637 A0A0T6B9V3 A0A1B6JIT1 A0A336KJ03 E9FUK4 A0A336MWP5 A0A1B0CUP8 A0A1L8DGC2 A0A1W4WME0 A0A1W4WCB4 A0A1B6GUL2 A0A182J920 A0A1L8DHG5 A0A2R7W5A6 A0A1I8NXM4 A0A1Y1NBJ8 A0A0A1XCZ8 A0A034VL94 W8BJF4 A0A0M4E8W2 A0A0K8U5N0 A0A1B6KX48 B4HYA3 A0A0J9QXY7 B4Q5T4 B4NZG9 B4KIM6 Q9VLX2 B3MV21 A0A1I8MSX1 B3N6N7 A0A2H8TDC1 A0A1W4W747 B4N0U7 B4JQD2 A0A0P5PWY6 A0A0P5K0M7
A0A0N1IPM5 A0A0L7L4J8 A0A0C9RUM2 A0A0C9RDX9 E2BEL8 A0A151WUU2 F4WAC0 A0A195CSG6 A0A158NKY0 A0A195FSA4 A0A195E454 E0VMG6 A0A154P2U9 A0A088A0B3 A0A2A3ES74 E9IZ18 A0A0J7L877 D2A3W1 A0A2J7RIK3 E2AL99 A0A067RX80 A0A026WME5 U5EPR5 A0A182HCX5 A0A1B0DRE2 A0A1B0DFM0 A0A0L7R1P3 A0A1Q3F2B7 A0A0A9Z2Y2 A0A0M9A3N5 A0A0K8SQJ5 A0A0A9Z6V7 A0A1B6DLS8 A0A182RAW2 A0A1Q3F3E8 A0A182Q0T5 A0A182P9G8 T1HMJ5 A0A182MZ24 A0A084W1X9 B0W8Y1 Q17J04 A0A1S4F196 A0A023EZU8 A0A0V0G908 W5J5K1 A0A069DXD8 A0A224XJV3 A0A182F637 A0A0T6B9V3 A0A1B6JIT1 A0A336KJ03 E9FUK4 A0A336MWP5 A0A1B0CUP8 A0A1L8DGC2 A0A1W4WME0 A0A1W4WCB4 A0A1B6GUL2 A0A182J920 A0A1L8DHG5 A0A2R7W5A6 A0A1I8NXM4 A0A1Y1NBJ8 A0A0A1XCZ8 A0A034VL94 W8BJF4 A0A0M4E8W2 A0A0K8U5N0 A0A1B6KX48 B4HYA3 A0A0J9QXY7 B4Q5T4 B4NZG9 B4KIM6 Q9VLX2 B3MV21 A0A1I8MSX1 B3N6N7 A0A2H8TDC1 A0A1W4W747 B4N0U7 B4JQD2 A0A0P5PWY6 A0A0P5K0M7
EC Number
2.1.2.1
Pubmed
19121390
28756777
23622113
22118469
26354079
26227816
+ More
20798317 21719571 21347285 20566863 21282665 18362917 19820115 24845553 24508170 30249741 26483478 25401762 26823975 24438588 17510324 25474469 20920257 23761445 26334808 21292972 28004739 25830018 25348373 24495485 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136
20798317 21719571 21347285 20566863 21282665 18362917 19820115 24845553 24508170 30249741 26483478 25401762 26823975 24438588 17510324 25474469 20920257 23761445 26334808 21292972 28004739 25830018 25348373 24495485 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136
EMBL
BABH01036188
ODYU01003736
SOQ42890.1
KZ150200
PZC72284.1
GAIX01003451
+ More
JAA89109.1 AGBW02008531 OWR53073.1 KQ459465 KPJ00345.1 KQ460401 KPJ14947.1 JTDY01003078 KOB70189.1 GBYB01011301 JAG81068.1 GBYB01011302 JAG81069.1 GL447829 EFN85852.1 KQ982718 KYQ51690.1 GL888048 EGI68803.1 KQ977306 KYN03653.1 ADTU01019014 ADTU01019015 KQ981281 KYN43321.1 KQ979701 KYN19687.1 DS235307 EEB14572.1 KQ434809 KZC06269.1 KZ288194 PBC33979.1 GL767078 EFZ14212.1 LBMM01000273 KMR02200.1 KQ971348 EFA04877.1 NEVH01003500 PNF40662.1 GL440553 EFN65795.1 KK852408 KDR24499.1 KK107152 QOIP01000007 EZA57195.1 RLU20067.1 GANO01003610 JAB56261.1 JXUM01033714 KQ560999 KXJ80081.1 AJVK01019958 AJVK01058828 KQ414667 KOC64785.1 GFDL01013368 JAV21677.1 GBHO01004012 GDHC01003561 JAG39592.1 JAQ15068.1 KQ435767 KOX75303.1 GBRD01010724 JAG55100.1 GBHO01004013 GDHC01017149 JAG39591.1 JAQ01480.1 GEDC01010669 JAS26629.1 GFDL01012988 JAV22057.1 AXCN02000792 ACPB03020506 ACPB03020507 ATLV01019471 KE525272 KFB44223.1 DS231860 EDS39259.1 CH477236 EAT46588.1 GBBI01003939 JAC14773.1 GECL01001521 JAP04603.1 ADMH02002094 ETN59266.1 GBGD01000547 JAC88342.1 GFTR01008003 JAW08423.1 LJIG01009051 KRT83853.1 GECU01008500 JAS99206.1 UFQS01000502 UFQT01000502 SSX04459.1 SSX24823.1 GL732525 EFX88744.1 UFQS01003099 UFQT01003099 SSX15195.1 SSX34570.1 AJWK01029522 AJWK01029523 AJWK01029524 AJWK01029525 AJWK01029526 AJWK01029527 AJWK01029528 AJWK01029529 AJWK01029530 GFDF01008581 JAV05503.1 GECZ01003647 JAS66122.1 GFDF01008257 JAV05827.1 KK854228 PTY13395.1 GEZM01007551 JAV95099.1 GBXI01005467 JAD08825.1 GAKP01014866 JAC44086.1 GAMC01007678 JAB98877.1 CP012523 ALC39440.1 GDHF01030347 JAI21967.1 GEBQ01023987 JAT15990.1 CH480818 EDW52033.1 CM002910 KMY88851.1 CM000361 EDX04130.1 KMY88850.1 CM000157 EDW87718.1 CH933807 EDW11369.1 AE014134 AY069504 AAF52557.1 AAL39649.1 CH902624 EDV33086.1 CH954177 EDV59253.1 GFXV01000302 MBW12107.1 CH963920 EDW77710.1 CH916372 EDV99112.1 GDIQ01132580 JAL19146.1 GDIQ01190894 JAK60831.1
JAA89109.1 AGBW02008531 OWR53073.1 KQ459465 KPJ00345.1 KQ460401 KPJ14947.1 JTDY01003078 KOB70189.1 GBYB01011301 JAG81068.1 GBYB01011302 JAG81069.1 GL447829 EFN85852.1 KQ982718 KYQ51690.1 GL888048 EGI68803.1 KQ977306 KYN03653.1 ADTU01019014 ADTU01019015 KQ981281 KYN43321.1 KQ979701 KYN19687.1 DS235307 EEB14572.1 KQ434809 KZC06269.1 KZ288194 PBC33979.1 GL767078 EFZ14212.1 LBMM01000273 KMR02200.1 KQ971348 EFA04877.1 NEVH01003500 PNF40662.1 GL440553 EFN65795.1 KK852408 KDR24499.1 KK107152 QOIP01000007 EZA57195.1 RLU20067.1 GANO01003610 JAB56261.1 JXUM01033714 KQ560999 KXJ80081.1 AJVK01019958 AJVK01058828 KQ414667 KOC64785.1 GFDL01013368 JAV21677.1 GBHO01004012 GDHC01003561 JAG39592.1 JAQ15068.1 KQ435767 KOX75303.1 GBRD01010724 JAG55100.1 GBHO01004013 GDHC01017149 JAG39591.1 JAQ01480.1 GEDC01010669 JAS26629.1 GFDL01012988 JAV22057.1 AXCN02000792 ACPB03020506 ACPB03020507 ATLV01019471 KE525272 KFB44223.1 DS231860 EDS39259.1 CH477236 EAT46588.1 GBBI01003939 JAC14773.1 GECL01001521 JAP04603.1 ADMH02002094 ETN59266.1 GBGD01000547 JAC88342.1 GFTR01008003 JAW08423.1 LJIG01009051 KRT83853.1 GECU01008500 JAS99206.1 UFQS01000502 UFQT01000502 SSX04459.1 SSX24823.1 GL732525 EFX88744.1 UFQS01003099 UFQT01003099 SSX15195.1 SSX34570.1 AJWK01029522 AJWK01029523 AJWK01029524 AJWK01029525 AJWK01029526 AJWK01029527 AJWK01029528 AJWK01029529 AJWK01029530 GFDF01008581 JAV05503.1 GECZ01003647 JAS66122.1 GFDF01008257 JAV05827.1 KK854228 PTY13395.1 GEZM01007551 JAV95099.1 GBXI01005467 JAD08825.1 GAKP01014866 JAC44086.1 GAMC01007678 JAB98877.1 CP012523 ALC39440.1 GDHF01030347 JAI21967.1 GEBQ01023987 JAT15990.1 CH480818 EDW52033.1 CM002910 KMY88851.1 CM000361 EDX04130.1 KMY88850.1 CM000157 EDW87718.1 CH933807 EDW11369.1 AE014134 AY069504 AAF52557.1 AAL39649.1 CH902624 EDV33086.1 CH954177 EDV59253.1 GFXV01000302 MBW12107.1 CH963920 EDW77710.1 CH916372 EDV99112.1 GDIQ01132580 JAL19146.1 GDIQ01190894 JAK60831.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000037510
UP000008237
+ More
UP000075809 UP000007755 UP000078542 UP000005205 UP000078541 UP000078492 UP000009046 UP000076502 UP000005203 UP000242457 UP000036403 UP000007266 UP000235965 UP000000311 UP000027135 UP000053097 UP000279307 UP000069940 UP000249989 UP000092462 UP000053825 UP000053105 UP000075900 UP000075886 UP000075885 UP000015103 UP000075884 UP000030765 UP000002320 UP000008820 UP000000673 UP000069272 UP000000305 UP000092461 UP000192223 UP000075880 UP000095300 UP000092553 UP000001292 UP000000304 UP000002282 UP000009192 UP000000803 UP000007801 UP000095301 UP000008711 UP000192221 UP000007798 UP000001070
UP000075809 UP000007755 UP000078542 UP000005205 UP000078541 UP000078492 UP000009046 UP000076502 UP000005203 UP000242457 UP000036403 UP000007266 UP000235965 UP000000311 UP000027135 UP000053097 UP000279307 UP000069940 UP000249989 UP000092462 UP000053825 UP000053105 UP000075900 UP000075886 UP000075885 UP000015103 UP000075884 UP000030765 UP000002320 UP000008820 UP000000673 UP000069272 UP000000305 UP000092461 UP000192223 UP000075880 UP000095300 UP000092553 UP000001292 UP000000304 UP000002282 UP000009192 UP000000803 UP000007801 UP000095301 UP000008711 UP000192221 UP000007798 UP000001070
Interpro
IPR036427
Bromodomain-like_sf
+ More
IPR021900 DUF3512
IPR001487 Bromodomain
IPR011990 TPR-like_helical_dom_sf
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR000210 BTB/POZ_dom
IPR001085 Ser_HO-MeTrfase
IPR019798 Ser_HO-MeTrfase_PLP_BS
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR013087 Znf_C2H2_type
IPR039429 SHMT-like_dom
IPR015421 PyrdxlP-dep_Trfase_major
IPR015424 PyrdxlP-dep_Trfase
IPR021900 DUF3512
IPR001487 Bromodomain
IPR011990 TPR-like_helical_dom_sf
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR000210 BTB/POZ_dom
IPR001085 Ser_HO-MeTrfase
IPR019798 Ser_HO-MeTrfase_PLP_BS
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR013087 Znf_C2H2_type
IPR039429 SHMT-like_dom
IPR015421 PyrdxlP-dep_Trfase_major
IPR015424 PyrdxlP-dep_Trfase
SUPFAM
Gene 3D
ProteinModelPortal
H9JVH5
A0A2H1VPZ8
A0A2W1BF18
S4PDX7
A0A212FH88
A0A194Q5R4
+ More
A0A0N1IPM5 A0A0L7L4J8 A0A0C9RUM2 A0A0C9RDX9 E2BEL8 A0A151WUU2 F4WAC0 A0A195CSG6 A0A158NKY0 A0A195FSA4 A0A195E454 E0VMG6 A0A154P2U9 A0A088A0B3 A0A2A3ES74 E9IZ18 A0A0J7L877 D2A3W1 A0A2J7RIK3 E2AL99 A0A067RX80 A0A026WME5 U5EPR5 A0A182HCX5 A0A1B0DRE2 A0A1B0DFM0 A0A0L7R1P3 A0A1Q3F2B7 A0A0A9Z2Y2 A0A0M9A3N5 A0A0K8SQJ5 A0A0A9Z6V7 A0A1B6DLS8 A0A182RAW2 A0A1Q3F3E8 A0A182Q0T5 A0A182P9G8 T1HMJ5 A0A182MZ24 A0A084W1X9 B0W8Y1 Q17J04 A0A1S4F196 A0A023EZU8 A0A0V0G908 W5J5K1 A0A069DXD8 A0A224XJV3 A0A182F637 A0A0T6B9V3 A0A1B6JIT1 A0A336KJ03 E9FUK4 A0A336MWP5 A0A1B0CUP8 A0A1L8DGC2 A0A1W4WME0 A0A1W4WCB4 A0A1B6GUL2 A0A182J920 A0A1L8DHG5 A0A2R7W5A6 A0A1I8NXM4 A0A1Y1NBJ8 A0A0A1XCZ8 A0A034VL94 W8BJF4 A0A0M4E8W2 A0A0K8U5N0 A0A1B6KX48 B4HYA3 A0A0J9QXY7 B4Q5T4 B4NZG9 B4KIM6 Q9VLX2 B3MV21 A0A1I8MSX1 B3N6N7 A0A2H8TDC1 A0A1W4W747 B4N0U7 B4JQD2 A0A0P5PWY6 A0A0P5K0M7
A0A0N1IPM5 A0A0L7L4J8 A0A0C9RUM2 A0A0C9RDX9 E2BEL8 A0A151WUU2 F4WAC0 A0A195CSG6 A0A158NKY0 A0A195FSA4 A0A195E454 E0VMG6 A0A154P2U9 A0A088A0B3 A0A2A3ES74 E9IZ18 A0A0J7L877 D2A3W1 A0A2J7RIK3 E2AL99 A0A067RX80 A0A026WME5 U5EPR5 A0A182HCX5 A0A1B0DRE2 A0A1B0DFM0 A0A0L7R1P3 A0A1Q3F2B7 A0A0A9Z2Y2 A0A0M9A3N5 A0A0K8SQJ5 A0A0A9Z6V7 A0A1B6DLS8 A0A182RAW2 A0A1Q3F3E8 A0A182Q0T5 A0A182P9G8 T1HMJ5 A0A182MZ24 A0A084W1X9 B0W8Y1 Q17J04 A0A1S4F196 A0A023EZU8 A0A0V0G908 W5J5K1 A0A069DXD8 A0A224XJV3 A0A182F637 A0A0T6B9V3 A0A1B6JIT1 A0A336KJ03 E9FUK4 A0A336MWP5 A0A1B0CUP8 A0A1L8DGC2 A0A1W4WME0 A0A1W4WCB4 A0A1B6GUL2 A0A182J920 A0A1L8DHG5 A0A2R7W5A6 A0A1I8NXM4 A0A1Y1NBJ8 A0A0A1XCZ8 A0A034VL94 W8BJF4 A0A0M4E8W2 A0A0K8U5N0 A0A1B6KX48 B4HYA3 A0A0J9QXY7 B4Q5T4 B4NZG9 B4KIM6 Q9VLX2 B3MV21 A0A1I8MSX1 B3N6N7 A0A2H8TDC1 A0A1W4W747 B4N0U7 B4JQD2 A0A0P5PWY6 A0A0P5K0M7
PDB
6HM0
E-value=1.29445e-26,
Score=298
Ontologies
GO
PANTHER
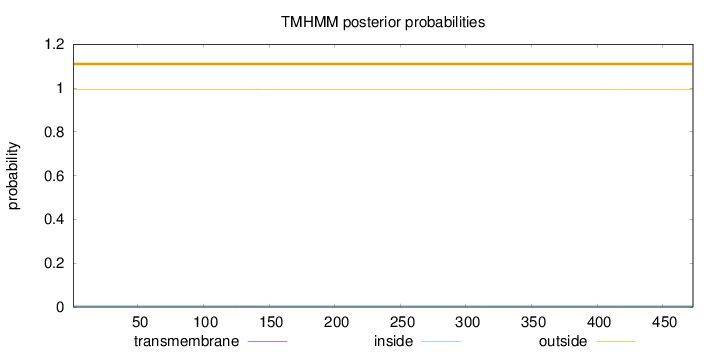
Topology
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00382
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00705
outside
1 - 473
Population Genetic Test Statistics
Pi
255.960538
Theta
187.855058
Tajima's D
1.27364
CLR
0.451407
CSRT
0.728763561821909
Interpretation
Uncertain
