Gene
KWMTBOMO02382 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013478
Annotation
HPO_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.796
Sequence
CDS
ATGGCGAAGCGCAACACGGTTATCGGCACCCCGTTTTGGATGGCGCCCGAGGTCATTCAGGAGATCGGGTACGACTGCGTGGCCGACATCTGGTCCCTGGGCATCACCGCTCTGGAGATGGCGGAGGGCAAGCCGCCCTACGGAGACATACATCCCATGAGGGCCATCTTCATGATACCCACCAAGCCGCCGCCTTCCTTCAGGGAGCCGGACCAGTGGTCGTCAGAGTTCATAGACTTCGTTAGTCAGTGCCTCATCAAGAACCCTGAAGACAGAGCCACAGCAGAGTTTCTATTGGCTCATGAGTTTATAGGTAACGCGAAGCAGCCCAGCATACTGAGCACCATGATAGCGGAGGCCCGGGAGCTGCGGGAGACGCAGCTGTACAGGAGCTCCCACCAGGCCTCCAACAACGCCAGGGCCTTTGCCGGGGGCCCCGACTACTCCGAGGCCACCATGAAGCAGCGCTCCGACGGGACGCTGGTGCCCGATCAGCTGGCCGACCTCAGCGCCGACCTGGGGACCATGGTCATCAACGAACCAGAACCGGACCTCGCCACCATGAAACGGCACGACACCGACCCCATGGACACCAACAAGCCGAAGTACCGGCCGCTGTTCTTGGAGCACTTTGACAAGAAGGAGATGAACCACTTGAACATCGGGGCCCCCACCAACGGGACCCTCACCGGCCCGCTGCGGCATCTTCCTCAGCACAGCATAGGTGGCGAGAGTGAGGCCAGCGAAGTGGGCGCGGCGGACATCCAGCGCGCGCTGATGGAGGAGGGGAACTTCGAATTCCTGTGGCAGCTGTCGGAGGCGGAGCTGGAGCAGCGCGTGGCGCGGCTGGACGCGCAGATGGAGCAGGAGATCGAGCTGCTGCGCGCGCGCTACGCTCGCAAGCGACAGCCCATCCTCGACGCCGTCACGCTCAAGCGGAAGCGACAGCAGAACTTCTAG
Protein
MAKRNTVIGTPFWMAPEVIQEIGYDCVADIWSLGITALEMAEGKPPYGDIHPMRAIFMIPTKPPPSFREPDQWSSEFIDFVSQCLIKNPEDRATAEFLLAHEFIGNAKQPSILSTMIAEARELRETQLYRSSHQASNNARAFAGGPDYSEATMKQRSDGTLVPDQLADLSADLGTMVINEPEPDLATMKRHDTDPMDTNKPKYRPLFLEHFDKKEMNHLNIGAPTNGTLTGPLRHLPQHSIGGESEASEVGAADIQRALMEEGNFEFLWQLSEAELEQRVARLDAQMEQEIELLRARYARKRQPILDAVTLKRKRQQNF
Summary
Uniprot
W6FFX1
B2CMB7
A0A2A4JTY8
A0A2W1BIV9
A0A2A4JT22
A0A2A4JTL1
+ More
A0A2A4JSZ4 A0A2A4JU64 A0A0K2BNK3 A0A0N0PCX1 A0A194Q5S0 A0A212ERT2 A0A2A4JUU0 A0A224XR53 A0A1B6H1B7 A0A1B6JJJ1 A0A1B6EEJ2 A0A023F5G2 A0A069DV68 A0A1B6C3S2 A0A0V0GCU6 A0A0N0BIQ5 A0A1B6LQ30 A0A0L7RIS0 A0A1B6MUF9 A0A088AHT1 A0A1W4WFI0 F4X6R2 A0A195E7M5 A0A195B2H0 A0A154P4U6 A0A195CI12 E2BVM0 A0A195F1Q0 D6W804 E0W4B5 A0A2S2QLP8 A0A2A3EA21 A0A158NFU0 A0A3L8D7A5 A0A0A9XY40 A0A2H8TEM1 J9K8L2 A0A0C9QSD3 K7J1R4 T1IK06 E9GI23 A0A0T6B942 A0A026WCQ3 A0A151XCE7 A0A0P5EUT7 A0A0P6EU47 A0A0P6HDB7 A0A0P5W8K0 A0A0N7ZEC7 A0A1Y1K4S0 N6U6J5 A0A182SL62 A0A0P5X941 T1HRI9 A0A0J7L993 A0A087TQG6 U4UVN1 A0A210PET8 A0A2R2MJQ1 K1PMC6 A0A0L8HLN0 A0A2P6KYG5 B7Q690 A0A293M530 A0A1B6E940 A0A084W434 A0A0B6ZA98 A0A0P5ZAW5 A0A2J7QKT4 V4B8W4 A0A2J7QKV9 A0A1Z5LGR8 E9IMK3 X5J6H2 A0A067REX7 A0A0K2SXQ6 A0A1S4FDS5 Q175W2 A0A182MX61 A0A1Y9IW72 A0A2H8TZX1 A0A2H8TR57 A0A182NKF3 A0A182GX42 A0A1Q3FHR5 A0A182YPM4 A0A182TRR8 A0A0P6DJM5 B0VZH7 A0A182FE28 A0A182KPB6 A0A0P5EE76 A0A2M4AP57
A0A2A4JSZ4 A0A2A4JU64 A0A0K2BNK3 A0A0N0PCX1 A0A194Q5S0 A0A212ERT2 A0A2A4JUU0 A0A224XR53 A0A1B6H1B7 A0A1B6JJJ1 A0A1B6EEJ2 A0A023F5G2 A0A069DV68 A0A1B6C3S2 A0A0V0GCU6 A0A0N0BIQ5 A0A1B6LQ30 A0A0L7RIS0 A0A1B6MUF9 A0A088AHT1 A0A1W4WFI0 F4X6R2 A0A195E7M5 A0A195B2H0 A0A154P4U6 A0A195CI12 E2BVM0 A0A195F1Q0 D6W804 E0W4B5 A0A2S2QLP8 A0A2A3EA21 A0A158NFU0 A0A3L8D7A5 A0A0A9XY40 A0A2H8TEM1 J9K8L2 A0A0C9QSD3 K7J1R4 T1IK06 E9GI23 A0A0T6B942 A0A026WCQ3 A0A151XCE7 A0A0P5EUT7 A0A0P6EU47 A0A0P6HDB7 A0A0P5W8K0 A0A0N7ZEC7 A0A1Y1K4S0 N6U6J5 A0A182SL62 A0A0P5X941 T1HRI9 A0A0J7L993 A0A087TQG6 U4UVN1 A0A210PET8 A0A2R2MJQ1 K1PMC6 A0A0L8HLN0 A0A2P6KYG5 B7Q690 A0A293M530 A0A1B6E940 A0A084W434 A0A0B6ZA98 A0A0P5ZAW5 A0A2J7QKT4 V4B8W4 A0A2J7QKV9 A0A1Z5LGR8 E9IMK3 X5J6H2 A0A067REX7 A0A0K2SXQ6 A0A1S4FDS5 Q175W2 A0A182MX61 A0A1Y9IW72 A0A2H8TZX1 A0A2H8TR57 A0A182NKF3 A0A182GX42 A0A1Q3FHR5 A0A182YPM4 A0A182TRR8 A0A0P6DJM5 B0VZH7 A0A182FE28 A0A182KPB6 A0A0P5EE76 A0A2M4AP57
Pubmed
28756777
26272745
26354079
22118469
25474469
26334808
+ More
21719571 20798317 18362917 19820115 20566863 21347285 30249741 25401762 26823975 20075255 21292972 24508170 28004739 23537049 28812685 26383154 22992520 24438588 23254933 28528879 21282665 23872316 24845553 17510324 26483478 25244985 20966253
21719571 20798317 18362917 19820115 20566863 21347285 30249741 25401762 26823975 20075255 21292972 24508170 28004739 23537049 28812685 26383154 22992520 24438588 23254933 28528879 21282665 23872316 24845553 17510324 26483478 25244985 20966253
EMBL
KF904333
AHJ25996.1
EU527369
ACB41090.1
NWSH01000630
PCG75158.1
+ More
KZ150157 PZC72766.1 PCG75161.1 PCG75159.1 PCG75165.1 PCG75164.1 KP735934 AKZ66620.1 KQ460401 KPJ14951.1 KQ459465 KPJ00350.1 AGBW02012923 OWR44210.1 PCG75162.1 GFTR01005506 JAW10920.1 GECZ01001300 JAS68469.1 GECU01020813 GECU01015155 GECU01008424 GECU01004514 GECU01003832 JAS86893.1 JAS92551.1 JAS99282.1 JAT03193.1 JAT03875.1 GEDC01000946 JAS36352.1 GBBI01002329 JAC16383.1 GBGD01001178 JAC87711.1 GEDC01029140 JAS08158.1 GECL01000935 JAP05189.1 KQ435727 KOX77898.1 GEBQ01014220 JAT25757.1 KQ414583 KOC70708.1 GEBQ01009209 GEBQ01000429 JAT30768.1 JAT39548.1 GL888818 EGI57844.1 KQ979568 KYN20829.1 KQ976662 KYM78487.1 KQ434806 KZC06150.1 KQ977754 KYN00067.1 GL450875 EFN80290.1 KQ981864 KYN34306.1 KQ971307 EFA11019.1 DS235886 EEB20471.1 GGMS01009474 MBY78677.1 KZ288310 PBC28575.1 ADTU01014740 QOIP01000012 RLU16340.1 GBHO01019821 GBRD01011262 GDHC01015342 JAG23783.1 JAG54562.1 JAQ03287.1 GFXV01000695 MBW12500.1 ABLF02030673 GBYB01003572 JAG73339.1 AFFK01014944 GL732545 EFX80950.1 LJIG01009286 KRT83383.1 KK107314 EZA52829.1 KQ982314 KYQ57968.1 GDIQ01269830 JAJ81894.1 GDIP01214120 GDIQ01209985 GDIQ01190857 GDIQ01101541 GDIQ01070851 LRGB01000868 JAJ09282.1 JAN23886.1 KZS15726.1 GDIQ01021139 JAN73598.1 GDIP01089706 JAM14009.1 GDIP01253721 JAI69680.1 GEZM01092715 JAV56489.1 APGK01047174 KB741077 ENN74177.1 GDIP01075592 JAM28123.1 ACPB03004826 LBMM01000226 KMR03928.1 KK116295 KFM67355.1 KB632384 ERL94311.1 NEDP02076746 OWF34966.1 JH815701 EKC20024.1 KQ417924 KOF89690.1 MWRG01003615 PRD31369.1 ABJB010302342 ABJB010418094 ABJB010501463 ABJB010677184 ABJB010911009 ABJB010998423 ABJB011061190 ABJB011071678 DS866317 EEC14349.1 GFWV01009346 MAA34075.1 GEDC01002862 JAS34436.1 ATLV01020271 KE525297 KFB44978.1 HACG01017775 CEK64640.1 GDIP01048638 JAM55077.1 NEVH01013256 PNF29197.1 KB203275 ESO85274.1 PNF29198.1 GFJQ02000849 JAW06121.1 GL764195 EFZ18201.1 HF969251 CCX34980.1 KK852549 KDR21583.1 HACA01001163 CDW18524.1 CH477395 EAT41841.1 AXCM01004994 GFXV01007664 MBW19469.1 GFXV01004277 MBW16082.1 JXUM01094626 JXUM01094627 KQ564141 KXJ72692.1 GFDL01007948 JAV27097.1 GDIQ01078002 JAN16735.1 DS231814 EDS31714.1 GDIP01144171 JAJ79231.1 GGFK01009077 MBW42398.1
KZ150157 PZC72766.1 PCG75161.1 PCG75159.1 PCG75165.1 PCG75164.1 KP735934 AKZ66620.1 KQ460401 KPJ14951.1 KQ459465 KPJ00350.1 AGBW02012923 OWR44210.1 PCG75162.1 GFTR01005506 JAW10920.1 GECZ01001300 JAS68469.1 GECU01020813 GECU01015155 GECU01008424 GECU01004514 GECU01003832 JAS86893.1 JAS92551.1 JAS99282.1 JAT03193.1 JAT03875.1 GEDC01000946 JAS36352.1 GBBI01002329 JAC16383.1 GBGD01001178 JAC87711.1 GEDC01029140 JAS08158.1 GECL01000935 JAP05189.1 KQ435727 KOX77898.1 GEBQ01014220 JAT25757.1 KQ414583 KOC70708.1 GEBQ01009209 GEBQ01000429 JAT30768.1 JAT39548.1 GL888818 EGI57844.1 KQ979568 KYN20829.1 KQ976662 KYM78487.1 KQ434806 KZC06150.1 KQ977754 KYN00067.1 GL450875 EFN80290.1 KQ981864 KYN34306.1 KQ971307 EFA11019.1 DS235886 EEB20471.1 GGMS01009474 MBY78677.1 KZ288310 PBC28575.1 ADTU01014740 QOIP01000012 RLU16340.1 GBHO01019821 GBRD01011262 GDHC01015342 JAG23783.1 JAG54562.1 JAQ03287.1 GFXV01000695 MBW12500.1 ABLF02030673 GBYB01003572 JAG73339.1 AFFK01014944 GL732545 EFX80950.1 LJIG01009286 KRT83383.1 KK107314 EZA52829.1 KQ982314 KYQ57968.1 GDIQ01269830 JAJ81894.1 GDIP01214120 GDIQ01209985 GDIQ01190857 GDIQ01101541 GDIQ01070851 LRGB01000868 JAJ09282.1 JAN23886.1 KZS15726.1 GDIQ01021139 JAN73598.1 GDIP01089706 JAM14009.1 GDIP01253721 JAI69680.1 GEZM01092715 JAV56489.1 APGK01047174 KB741077 ENN74177.1 GDIP01075592 JAM28123.1 ACPB03004826 LBMM01000226 KMR03928.1 KK116295 KFM67355.1 KB632384 ERL94311.1 NEDP02076746 OWF34966.1 JH815701 EKC20024.1 KQ417924 KOF89690.1 MWRG01003615 PRD31369.1 ABJB010302342 ABJB010418094 ABJB010501463 ABJB010677184 ABJB010911009 ABJB010998423 ABJB011061190 ABJB011071678 DS866317 EEC14349.1 GFWV01009346 MAA34075.1 GEDC01002862 JAS34436.1 ATLV01020271 KE525297 KFB44978.1 HACG01017775 CEK64640.1 GDIP01048638 JAM55077.1 NEVH01013256 PNF29197.1 KB203275 ESO85274.1 PNF29198.1 GFJQ02000849 JAW06121.1 GL764195 EFZ18201.1 HF969251 CCX34980.1 KK852549 KDR21583.1 HACA01001163 CDW18524.1 CH477395 EAT41841.1 AXCM01004994 GFXV01007664 MBW19469.1 GFXV01004277 MBW16082.1 JXUM01094626 JXUM01094627 KQ564141 KXJ72692.1 GFDL01007948 JAV27097.1 GDIQ01078002 JAN16735.1 DS231814 EDS31714.1 GDIP01144171 JAJ79231.1 GGFK01009077 MBW42398.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000053105
UP000053825
+ More
UP000005203 UP000192223 UP000007755 UP000078492 UP000078540 UP000076502 UP000078542 UP000008237 UP000078541 UP000007266 UP000009046 UP000242457 UP000005205 UP000279307 UP000007819 UP000002358 UP000000305 UP000053097 UP000075809 UP000076858 UP000019118 UP000075901 UP000015103 UP000036403 UP000054359 UP000030742 UP000242188 UP000085678 UP000005408 UP000053454 UP000001555 UP000030765 UP000235965 UP000030746 UP000027135 UP000008820 UP000075883 UP000075920 UP000075884 UP000069940 UP000249989 UP000076408 UP000075902 UP000002320 UP000069272 UP000075882
UP000005203 UP000192223 UP000007755 UP000078492 UP000078540 UP000076502 UP000078542 UP000008237 UP000078541 UP000007266 UP000009046 UP000242457 UP000005205 UP000279307 UP000007819 UP000002358 UP000000305 UP000053097 UP000075809 UP000076858 UP000019118 UP000075901 UP000015103 UP000036403 UP000054359 UP000030742 UP000242188 UP000085678 UP000005408 UP000053454 UP000001555 UP000030765 UP000235965 UP000030746 UP000027135 UP000008820 UP000075883 UP000075920 UP000075884 UP000069940 UP000249989 UP000076408 UP000075902 UP000002320 UP000069272 UP000075882
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
W6FFX1
B2CMB7
A0A2A4JTY8
A0A2W1BIV9
A0A2A4JT22
A0A2A4JTL1
+ More
A0A2A4JSZ4 A0A2A4JU64 A0A0K2BNK3 A0A0N0PCX1 A0A194Q5S0 A0A212ERT2 A0A2A4JUU0 A0A224XR53 A0A1B6H1B7 A0A1B6JJJ1 A0A1B6EEJ2 A0A023F5G2 A0A069DV68 A0A1B6C3S2 A0A0V0GCU6 A0A0N0BIQ5 A0A1B6LQ30 A0A0L7RIS0 A0A1B6MUF9 A0A088AHT1 A0A1W4WFI0 F4X6R2 A0A195E7M5 A0A195B2H0 A0A154P4U6 A0A195CI12 E2BVM0 A0A195F1Q0 D6W804 E0W4B5 A0A2S2QLP8 A0A2A3EA21 A0A158NFU0 A0A3L8D7A5 A0A0A9XY40 A0A2H8TEM1 J9K8L2 A0A0C9QSD3 K7J1R4 T1IK06 E9GI23 A0A0T6B942 A0A026WCQ3 A0A151XCE7 A0A0P5EUT7 A0A0P6EU47 A0A0P6HDB7 A0A0P5W8K0 A0A0N7ZEC7 A0A1Y1K4S0 N6U6J5 A0A182SL62 A0A0P5X941 T1HRI9 A0A0J7L993 A0A087TQG6 U4UVN1 A0A210PET8 A0A2R2MJQ1 K1PMC6 A0A0L8HLN0 A0A2P6KYG5 B7Q690 A0A293M530 A0A1B6E940 A0A084W434 A0A0B6ZA98 A0A0P5ZAW5 A0A2J7QKT4 V4B8W4 A0A2J7QKV9 A0A1Z5LGR8 E9IMK3 X5J6H2 A0A067REX7 A0A0K2SXQ6 A0A1S4FDS5 Q175W2 A0A182MX61 A0A1Y9IW72 A0A2H8TZX1 A0A2H8TR57 A0A182NKF3 A0A182GX42 A0A1Q3FHR5 A0A182YPM4 A0A182TRR8 A0A0P6DJM5 B0VZH7 A0A182FE28 A0A182KPB6 A0A0P5EE76 A0A2M4AP57
A0A2A4JSZ4 A0A2A4JU64 A0A0K2BNK3 A0A0N0PCX1 A0A194Q5S0 A0A212ERT2 A0A2A4JUU0 A0A224XR53 A0A1B6H1B7 A0A1B6JJJ1 A0A1B6EEJ2 A0A023F5G2 A0A069DV68 A0A1B6C3S2 A0A0V0GCU6 A0A0N0BIQ5 A0A1B6LQ30 A0A0L7RIS0 A0A1B6MUF9 A0A088AHT1 A0A1W4WFI0 F4X6R2 A0A195E7M5 A0A195B2H0 A0A154P4U6 A0A195CI12 E2BVM0 A0A195F1Q0 D6W804 E0W4B5 A0A2S2QLP8 A0A2A3EA21 A0A158NFU0 A0A3L8D7A5 A0A0A9XY40 A0A2H8TEM1 J9K8L2 A0A0C9QSD3 K7J1R4 T1IK06 E9GI23 A0A0T6B942 A0A026WCQ3 A0A151XCE7 A0A0P5EUT7 A0A0P6EU47 A0A0P6HDB7 A0A0P5W8K0 A0A0N7ZEC7 A0A1Y1K4S0 N6U6J5 A0A182SL62 A0A0P5X941 T1HRI9 A0A0J7L993 A0A087TQG6 U4UVN1 A0A210PET8 A0A2R2MJQ1 K1PMC6 A0A0L8HLN0 A0A2P6KYG5 B7Q690 A0A293M530 A0A1B6E940 A0A084W434 A0A0B6ZA98 A0A0P5ZAW5 A0A2J7QKT4 V4B8W4 A0A2J7QKV9 A0A1Z5LGR8 E9IMK3 X5J6H2 A0A067REX7 A0A0K2SXQ6 A0A1S4FDS5 Q175W2 A0A182MX61 A0A1Y9IW72 A0A2H8TZX1 A0A2H8TR57 A0A182NKF3 A0A182GX42 A0A1Q3FHR5 A0A182YPM4 A0A182TRR8 A0A0P6DJM5 B0VZH7 A0A182FE28 A0A182KPB6 A0A0P5EE76 A0A2M4AP57
PDB
4LG4
E-value=7.34553e-54,
Score=531
Ontologies
KEGG
PATHWAY
GO
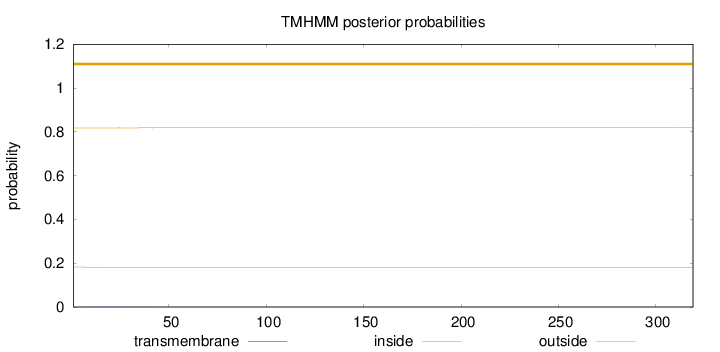
Topology
Length:
319
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01337
Exp number, first 60 AAs:
0.01314
Total prob of N-in:
0.18223
outside
1 - 319
Population Genetic Test Statistics
Pi
244.977425
Theta
205.420276
Tajima's D
0.494468
CLR
0.194292
CSRT
0.510274486275686
Interpretation
Uncertain
