Pre Gene Modal
BGIBMGA013543
Annotation
Transcription_factor_2B_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 1.36 Nuclear Reliability : 1.134
Sequence
CDS
ATGCCCATGAGGTTGGACGAAGGCTGGAACCAGATACAGTTCAACTTGGCTGACTTCACACGAAGAGCTTACGGCACTAACTATGTTGAGACCTTAAGAGTCCAAATACATGCAAACTGCAGAATAAGAAGAGTTTATTTCTCCGACAGATTGTATTCAGAAGATGAGCTCCCAGCAGAGTTCAAATTGTTCCTTCCAATTCAAAATAAAGCCAAAGTAGCAGCAGCAACTTAA
Protein
MPMRLDEGWNQIQFNLADFTRRAYGTNYVETLRVQIHANCRIRRVYFSDRLYSEDELPAEFKLFLPIQNKAKVAAAT
Summary
Uniprot
F6QRE2
A0A3B3Q5M1
A0A0P7Z0P2
H9JVI0
A0A0L7LMB6
S4NSG9
+ More
A0A212ERX0 A0A2W1BGC9 A0A0N1IA89 A0A194QAB1 A0A2H1W005 A0A1E1W055 A0A2A4JSZ2 Q1HQ38 J3JTF3 A0A1Y1KEP5 A0A1B6M2M2 A0A232EK26 A0A195CLI5 A0A195E4T3 A0A0L7RIJ9 F4W479 A0A195B3H1 K7IQR7 A0A2A3EED1 A0A088ARI1 A0A195ET87 E2B6M8 A0A158NFG8 E2A716 A0A151X287 A0A026VW04 E9IE47 A0A3B3Q5N5 A0A2P8XM97 A0A2J7QA74 V8NUH9 A0A3P9AL65 A0A1W4YGL5 A0A1W4Y7D9 G3VRK9 U3I0D9 H0Z2V2 A0A3M0KQB5 A0A093BWW8 W5MLJ3 A0A067RA15 A0A2U3ZB82 A0A3B1KG92 E0W4A2 A0A2B4SY99 A0A0P5FRT9 M3WMG3 G1MFS2 G3TMW6 F7C1L3 F6R525 H0WMG0 C3ZB94 A0A1B6CN42 A0A182GKY0 A0A293MLL5 A0A1S3CXK6 A0A1B6GCR3 A0A1B6JTP1 K9IGE5 A0A2U3XNG8 A0A0P5ZNY1 G1MX21 A0A0V0GAM3 A0A069DP20 A0A0A9Y236 A0A087V3Q3 A0A094NHT6 G5AUG3 D2GTY5 A0A093CAF2 A0A093IG56 A0A099ZBP3 A0A093F5R4 A0A093SEL8 A0A091U2H8 A0A091VKH0 A0A093TF24 A0A091XVZ3 R0LKP8 A0A091JIU8 A0A0A0A2D9 A0A093BQ61 A0A091PA40 A0A093NHD3 A0A091S1W9 A0A093HQ40 A0A091KLU6 A0A091F182 A0A093JBX8 A0A091R749 A0A091SLR0 A0A091PTH9 A0A087QN60
A0A212ERX0 A0A2W1BGC9 A0A0N1IA89 A0A194QAB1 A0A2H1W005 A0A1E1W055 A0A2A4JSZ2 Q1HQ38 J3JTF3 A0A1Y1KEP5 A0A1B6M2M2 A0A232EK26 A0A195CLI5 A0A195E4T3 A0A0L7RIJ9 F4W479 A0A195B3H1 K7IQR7 A0A2A3EED1 A0A088ARI1 A0A195ET87 E2B6M8 A0A158NFG8 E2A716 A0A151X287 A0A026VW04 E9IE47 A0A3B3Q5N5 A0A2P8XM97 A0A2J7QA74 V8NUH9 A0A3P9AL65 A0A1W4YGL5 A0A1W4Y7D9 G3VRK9 U3I0D9 H0Z2V2 A0A3M0KQB5 A0A093BWW8 W5MLJ3 A0A067RA15 A0A2U3ZB82 A0A3B1KG92 E0W4A2 A0A2B4SY99 A0A0P5FRT9 M3WMG3 G1MFS2 G3TMW6 F7C1L3 F6R525 H0WMG0 C3ZB94 A0A1B6CN42 A0A182GKY0 A0A293MLL5 A0A1S3CXK6 A0A1B6GCR3 A0A1B6JTP1 K9IGE5 A0A2U3XNG8 A0A0P5ZNY1 G1MX21 A0A0V0GAM3 A0A069DP20 A0A0A9Y236 A0A087V3Q3 A0A094NHT6 G5AUG3 D2GTY5 A0A093CAF2 A0A093IG56 A0A099ZBP3 A0A093F5R4 A0A093SEL8 A0A091U2H8 A0A091VKH0 A0A093TF24 A0A091XVZ3 R0LKP8 A0A091JIU8 A0A0A0A2D9 A0A093BQ61 A0A091PA40 A0A093NHD3 A0A091S1W9 A0A093HQ40 A0A091KLU6 A0A091F182 A0A093JBX8 A0A091R749 A0A091SLR0 A0A091PTH9 A0A087QN60
Pubmed
29240929
19121390
26227816
23622113
22118469
28756777
+ More
26354079 22516182 23537049 28004739 28648823 21719571 20075255 20798317 21347285 24508170 30249741 21282665 29403074 24297900 25069045 21709235 23749191 20360741 24845553 25329095 20566863 17975172 20010809 19892987 17495919 18563158 26483478 20838655 26334808 25401762 26823975 21993625
26354079 22516182 23537049 28004739 28648823 21719571 20075255 20798317 21347285 24508170 30249741 21282665 29403074 24297900 25069045 21709235 23749191 20360741 24845553 25329095 20566863 17975172 20010809 19892987 17495919 18563158 26483478 20838655 26334808 25401762 26823975 21993625
EMBL
JARO02002006
KPP73850.1
BABH01040483
JTDY01000581
KOB76587.1
GAIX01014102
+ More
JAA78458.1 AGBW02012923 OWR44209.1 KZ150157 PZC72765.1 KQ460401 KPJ14953.1 KQ459465 KPJ00351.1 ODYU01005207 SOQ45834.1 GDQN01010735 JAT80319.1 NWSH01000630 PCG75157.1 DQ443214 ABF51303.1 APGK01042151 BT126509 KB741002 KB631903 AEE61473.1 ENN75735.1 ERL87024.1 GEZM01088579 JAV58085.1 GEBQ01009829 JAT30148.1 NNAY01003892 OXU18707.1 KQ977642 KYN00934.1 KQ979609 KYN20173.1 KQ414584 KOC70628.1 GL887491 EGI71075.1 KQ976625 KYM79036.1 KZ288282 PBC29579.1 KQ981979 KYN31465.1 GL446000 EFN88612.1 ADTU01001532 GL437252 EFN70748.1 KQ982585 KYQ54304.1 KK107796 QOIP01000013 EZA47671.1 RLU15597.1 GL762556 EFZ21157.1 PYGN01001738 PSN33084.1 NEVH01016332 PNF25485.1 AZIM01001962 ETE65197.1 AEFK01035029 ADON01075948 ABQF01124045 QRBI01000116 RMC09237.1 KL454467 KFV08195.1 AHAT01018159 KK852605 KDR20427.1 DS235886 EEB20458.1 LSMT01000013 PFX33345.1 GDIQ01254158 JAJ97566.1 AANG04002512 ACTA01056789 AAQR03092967 AAQR03092968 GG666603 EEN50131.1 GEDC01022475 JAS14823.1 JXUM01071301 KQ562655 KXJ75391.1 GFWV01016648 MAA41376.1 GECZ01009558 JAS60211.1 GECU01005177 JAT02530.1 GABZ01008080 JAA45445.1 GDIP01042076 JAM61639.1 GECL01001666 JAP04458.1 GBGD01003076 JAC85813.1 GBHO01020019 GDHC01008890 JAG23585.1 JAQ09739.1 KL478745 KFO07245.1 KL275395 KFZ66046.1 JH167030 EHB00674.1 GL192338 EFB22241.1 KL236623 KFV09292.1 KK591706 KFW00598.1 KL891798 KGL79162.1 KK617714 KFV49546.1 KL671099 KFW81179.1 KK414817 KFQ84979.1 KL411055 KFR02938.1 KL436330 KFW92979.1 KK736075 KFR17066.1 KB743139 EOB00993.1 KK502030 KFP19705.1 KL870331 KGL87563.1 KN127047 KFU94724.1 KK670712 KFQ04787.1 KL224770 KFW63556.1 KK938143 KFQ48539.1 KK394829 KFV56673.1 KK745699 KFP40155.1 KK719362 KFO63016.1 KL205902 KFV76379.1 KK810335 KFQ35082.1 KK477974 KFQ59602.1 KK665399 KFQ10970.1 KL225750 KFM02664.1
JAA78458.1 AGBW02012923 OWR44209.1 KZ150157 PZC72765.1 KQ460401 KPJ14953.1 KQ459465 KPJ00351.1 ODYU01005207 SOQ45834.1 GDQN01010735 JAT80319.1 NWSH01000630 PCG75157.1 DQ443214 ABF51303.1 APGK01042151 BT126509 KB741002 KB631903 AEE61473.1 ENN75735.1 ERL87024.1 GEZM01088579 JAV58085.1 GEBQ01009829 JAT30148.1 NNAY01003892 OXU18707.1 KQ977642 KYN00934.1 KQ979609 KYN20173.1 KQ414584 KOC70628.1 GL887491 EGI71075.1 KQ976625 KYM79036.1 KZ288282 PBC29579.1 KQ981979 KYN31465.1 GL446000 EFN88612.1 ADTU01001532 GL437252 EFN70748.1 KQ982585 KYQ54304.1 KK107796 QOIP01000013 EZA47671.1 RLU15597.1 GL762556 EFZ21157.1 PYGN01001738 PSN33084.1 NEVH01016332 PNF25485.1 AZIM01001962 ETE65197.1 AEFK01035029 ADON01075948 ABQF01124045 QRBI01000116 RMC09237.1 KL454467 KFV08195.1 AHAT01018159 KK852605 KDR20427.1 DS235886 EEB20458.1 LSMT01000013 PFX33345.1 GDIQ01254158 JAJ97566.1 AANG04002512 ACTA01056789 AAQR03092967 AAQR03092968 GG666603 EEN50131.1 GEDC01022475 JAS14823.1 JXUM01071301 KQ562655 KXJ75391.1 GFWV01016648 MAA41376.1 GECZ01009558 JAS60211.1 GECU01005177 JAT02530.1 GABZ01008080 JAA45445.1 GDIP01042076 JAM61639.1 GECL01001666 JAP04458.1 GBGD01003076 JAC85813.1 GBHO01020019 GDHC01008890 JAG23585.1 JAQ09739.1 KL478745 KFO07245.1 KL275395 KFZ66046.1 JH167030 EHB00674.1 GL192338 EFB22241.1 KL236623 KFV09292.1 KK591706 KFW00598.1 KL891798 KGL79162.1 KK617714 KFV49546.1 KL671099 KFW81179.1 KK414817 KFQ84979.1 KL411055 KFR02938.1 KL436330 KFW92979.1 KK736075 KFR17066.1 KB743139 EOB00993.1 KK502030 KFP19705.1 KL870331 KGL87563.1 KN127047 KFU94724.1 KK670712 KFQ04787.1 KL224770 KFW63556.1 KK938143 KFQ48539.1 KK394829 KFV56673.1 KK745699 KFP40155.1 KK719362 KFO63016.1 KL205902 KFV76379.1 KK810335 KFQ35082.1 KK477974 KFQ59602.1 KK665399 KFQ10970.1 KL225750 KFM02664.1
Proteomes
UP000008225
UP000261540
UP000034805
UP000005204
UP000037510
UP000007151
+ More
UP000053240 UP000053268 UP000218220 UP000019118 UP000030742 UP000215335 UP000078542 UP000078492 UP000053825 UP000007755 UP000078540 UP000002358 UP000242457 UP000005203 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000245037 UP000235965 UP000265140 UP000192224 UP000007648 UP000016666 UP000007754 UP000269221 UP000018468 UP000027135 UP000245340 UP000018467 UP000009046 UP000225706 UP000011712 UP000008912 UP000007646 UP000002281 UP000002280 UP000005225 UP000001554 UP000069940 UP000249989 UP000079169 UP000245341 UP000001645 UP000006813 UP000053641 UP000053258 UP000053283 UP000053605 UP000053119 UP000053858 UP000054081 UP000052976 UP000053584 UP000053286
UP000053240 UP000053268 UP000218220 UP000019118 UP000030742 UP000215335 UP000078542 UP000078492 UP000053825 UP000007755 UP000078540 UP000002358 UP000242457 UP000005203 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000245037 UP000235965 UP000265140 UP000192224 UP000007648 UP000016666 UP000007754 UP000269221 UP000018468 UP000027135 UP000245340 UP000018467 UP000009046 UP000225706 UP000011712 UP000008912 UP000007646 UP000002281 UP000002280 UP000005225 UP000001554 UP000069940 UP000249989 UP000079169 UP000245341 UP000001645 UP000006813 UP000053641 UP000053258 UP000053283 UP000053605 UP000053119 UP000053858 UP000054081 UP000052976 UP000053584 UP000053286
Pfam
PF05018 DUF667
ProteinModelPortal
F6QRE2
A0A3B3Q5M1
A0A0P7Z0P2
H9JVI0
A0A0L7LMB6
S4NSG9
+ More
A0A212ERX0 A0A2W1BGC9 A0A0N1IA89 A0A194QAB1 A0A2H1W005 A0A1E1W055 A0A2A4JSZ2 Q1HQ38 J3JTF3 A0A1Y1KEP5 A0A1B6M2M2 A0A232EK26 A0A195CLI5 A0A195E4T3 A0A0L7RIJ9 F4W479 A0A195B3H1 K7IQR7 A0A2A3EED1 A0A088ARI1 A0A195ET87 E2B6M8 A0A158NFG8 E2A716 A0A151X287 A0A026VW04 E9IE47 A0A3B3Q5N5 A0A2P8XM97 A0A2J7QA74 V8NUH9 A0A3P9AL65 A0A1W4YGL5 A0A1W4Y7D9 G3VRK9 U3I0D9 H0Z2V2 A0A3M0KQB5 A0A093BWW8 W5MLJ3 A0A067RA15 A0A2U3ZB82 A0A3B1KG92 E0W4A2 A0A2B4SY99 A0A0P5FRT9 M3WMG3 G1MFS2 G3TMW6 F7C1L3 F6R525 H0WMG0 C3ZB94 A0A1B6CN42 A0A182GKY0 A0A293MLL5 A0A1S3CXK6 A0A1B6GCR3 A0A1B6JTP1 K9IGE5 A0A2U3XNG8 A0A0P5ZNY1 G1MX21 A0A0V0GAM3 A0A069DP20 A0A0A9Y236 A0A087V3Q3 A0A094NHT6 G5AUG3 D2GTY5 A0A093CAF2 A0A093IG56 A0A099ZBP3 A0A093F5R4 A0A093SEL8 A0A091U2H8 A0A091VKH0 A0A093TF24 A0A091XVZ3 R0LKP8 A0A091JIU8 A0A0A0A2D9 A0A093BQ61 A0A091PA40 A0A093NHD3 A0A091S1W9 A0A093HQ40 A0A091KLU6 A0A091F182 A0A093JBX8 A0A091R749 A0A091SLR0 A0A091PTH9 A0A087QN60
A0A212ERX0 A0A2W1BGC9 A0A0N1IA89 A0A194QAB1 A0A2H1W005 A0A1E1W055 A0A2A4JSZ2 Q1HQ38 J3JTF3 A0A1Y1KEP5 A0A1B6M2M2 A0A232EK26 A0A195CLI5 A0A195E4T3 A0A0L7RIJ9 F4W479 A0A195B3H1 K7IQR7 A0A2A3EED1 A0A088ARI1 A0A195ET87 E2B6M8 A0A158NFG8 E2A716 A0A151X287 A0A026VW04 E9IE47 A0A3B3Q5N5 A0A2P8XM97 A0A2J7QA74 V8NUH9 A0A3P9AL65 A0A1W4YGL5 A0A1W4Y7D9 G3VRK9 U3I0D9 H0Z2V2 A0A3M0KQB5 A0A093BWW8 W5MLJ3 A0A067RA15 A0A2U3ZB82 A0A3B1KG92 E0W4A2 A0A2B4SY99 A0A0P5FRT9 M3WMG3 G1MFS2 G3TMW6 F7C1L3 F6R525 H0WMG0 C3ZB94 A0A1B6CN42 A0A182GKY0 A0A293MLL5 A0A1S3CXK6 A0A1B6GCR3 A0A1B6JTP1 K9IGE5 A0A2U3XNG8 A0A0P5ZNY1 G1MX21 A0A0V0GAM3 A0A069DP20 A0A0A9Y236 A0A087V3Q3 A0A094NHT6 G5AUG3 D2GTY5 A0A093CAF2 A0A093IG56 A0A099ZBP3 A0A093F5R4 A0A093SEL8 A0A091U2H8 A0A091VKH0 A0A093TF24 A0A091XVZ3 R0LKP8 A0A091JIU8 A0A0A0A2D9 A0A093BQ61 A0A091PA40 A0A093NHD3 A0A091S1W9 A0A093HQ40 A0A091KLU6 A0A091F182 A0A093JBX8 A0A091R749 A0A091SLR0 A0A091PTH9 A0A087QN60
Ontologies
GO
PANTHER
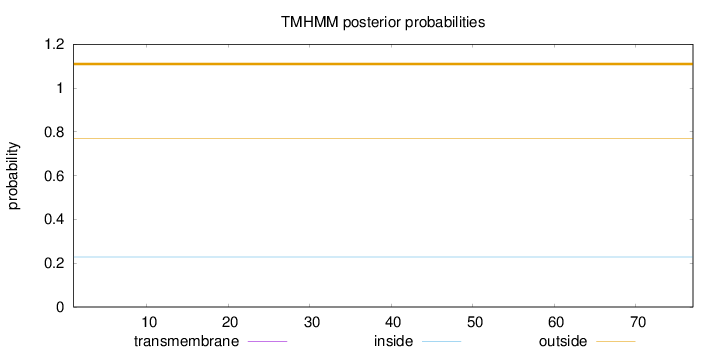
Topology
Length:
77
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00055
Exp number, first 60 AAs:
0.00055
Total prob of N-in:
0.22887
outside
1 - 77
Population Genetic Test Statistics
Pi
183.176569
Theta
186.601321
Tajima's D
-0.122568
CLR
40.352104
CSRT
0.332783360831958
Interpretation
Uncertain
